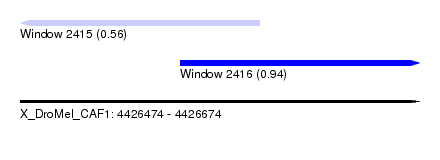
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,426,474 – 4,426,674 |
| Length | 200 |
| Max. P | 0.942429 |

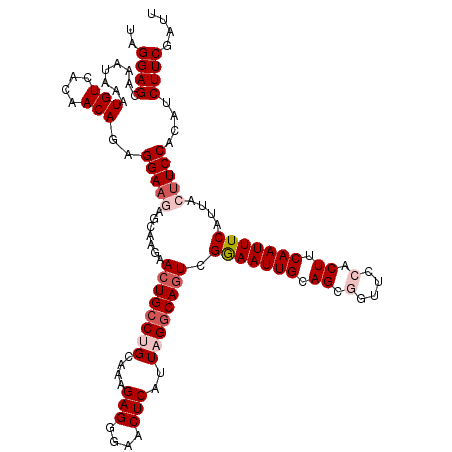
| Location | 4,426,474 – 4,426,594 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.64 |
| Mean single sequence MFE | -32.16 |
| Consensus MFE | -21.86 |
| Energy contribution | -23.34 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.559561 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4426474 120 - 22224390 GUUGUUGCUCUUCCUCUGUUGUGACAUUUAUUUGCUCCUAAUUCCUGCGGUUAUCUCCGCCAGAAGGUUAGCCAGAGAUAGUCCAAUUAUGCAUGGAUUGUCCAGGAAGAGUCAUCAUCC ..((.(((((((((((((..(..(.......)..)..((((((.((((((......))).)))..)))))).))))(((((((((........)))))))))..))))))).)).))... ( -35.50) >DroSec_CAF1 28856 120 - 1 GUUCUUGCUCUUCCUCUGUUGUGACAUUUAUUUGCUCCUAAUUCCCGUGGUUAUCUUCGCCAGAAGGUUAGCCAGAGAUAGUCCAAUUAUGCAUGGAUUGUCCAGGAGGAGUCAUCAUCC ......(((((((((..((((.((((......)).)).)))).....(((((((((((....))))).))))))(.(((((((((........)))))))))))))))))))........ ( -34.90) >DroSim_CAF1 30174 108 - 1 GUUCUUGCUCUUCCUCUGUUGUGACAUUUAUUUGCUCCUAAUUCCUGCGGUUAUCUCCGCC------------AGAGAUAGUCCAAUUAUGCAUGGAUUGUCCAGGAGGAGUCAUCAUCC ......(((((((((((((((.((((......)).)).))).....((((......)))))------------)))(((((((((........)))))))))..))))))))........ ( -31.30) >DroEre_CAF1 29802 117 - 1 GUUAUU---UUUCCUCUGUUGUGACAUUUAUUUGCUCCUAAUUCCUGUGGUUAUCUCCGCCAGAAGGUUAGCCAAAGAUAGUCCAAUUAUGCAUGGAUUGUCCAGGAAGAGUCAUCAUCC (((((.---.(......)..)))))........((((....((((((((((((...((.......)).))))))..(((((((((........)))))))))))))))))))........ ( -30.20) >DroYak_CAF1 29414 117 - 1 GUUAUU---UGUCCUCUGUUAUGACAAAUAUUUCCUCCUAAUUCCUGUGGUUAUCUCCGUCAGAAGGUUAGCCAGAGAUUGUCCAAUUAUGUAUGGAUUGUCCAGGAAGAGUCAUCAUCC ..((((---((((.........))))))))....(((....((((((((((((...((.......)).))))))..(((.(((((........))))).))))))))))))......... ( -28.90) >consensus GUUAUUGCUCUUCCUCUGUUGUGACAUUUAUUUGCUCCUAAUUCCUGUGGUUAUCUCCGCCAGAAGGUUAGCCAGAGAUAGUCCAAUUAUGCAUGGAUUGUCCAGGAAGAGUCAUCAUCC ......(((((((((...............((((.......(((..((((......))))..))).......))))(((((((((........))))))))).)))))))))........ (-21.86 = -23.34 + 1.48)

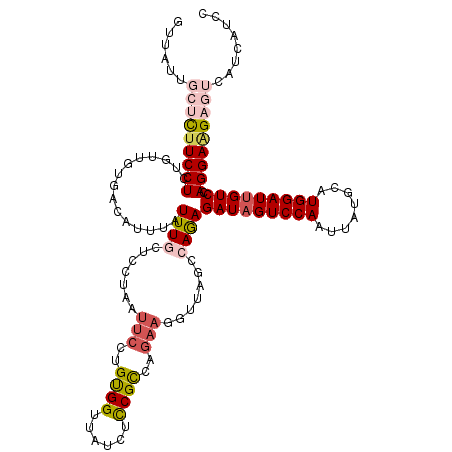
| Location | 4,426,554 – 4,426,674 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.97 |
| Mean single sequence MFE | -29.04 |
| Consensus MFE | -25.08 |
| Energy contribution | -26.64 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.942429 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4426554 120 + 22224390 UAGGAGCAAAUAAAUGUCACAACAGAGGAAGAGCAACAACUGCCUGCAAAGAGGGAACUCAUUAGUCAGUCGGAAUUGCAGCGGUUCCACUUCAAUUUCAUUACUUCCACAUCUUCGAUU ..((((........(((....)))..(((((.......((((.(((....(((....)))..))).)))).(((((((.((.(....).)).)))))))....)))))....)))).... ( -26.40) >DroSec_CAF1 28936 120 + 1 UAGGAGCAAAUAAAUGUCACAACAGAGGAAGAGCAAGAACUGCCAGCAAAGAGGGAACUCAUUAGGCAGUCGAAAUUGCAGCGGUUCCACUUCAAUUUCAUUACUUCCACAUCUUCGAUU ..((((........(((....)))..(((((.....((.(((((......(((....)))....)))))))(((((((.((.(....).)).)))))))....)))))....)))).... ( -31.70) >DroSim_CAF1 30242 120 + 1 UAGGAGCAAAUAAAUGUCACAACAGAGGAAGAGCAAGAACUGCCAGCAAAGAGGGAACUCAUUAGGCAGUCGAAAUUGCAGCGGUUCCACUUCAAUUUCAUUACUUCCACAUCUUCGAUU ..((((........(((....)))..(((((.....((.(((((......(((....)))....)))))))(((((((.((.(....).)).)))))))....)))))....)))).... ( -31.70) >DroEre_CAF1 29882 116 + 1 UAGGAGCAAAUAAAUGUCACAACAGAGGAAA---AAUAACUGCCUGCAAGGAGG-CACUCAUUAGGCAAUCGGAAUUGCAGCAGUUCCACUUCAAUUUCAUUACUUCCACAUCUUCGAUU ..((((........(((....)))((((...---......((((((....(((.-..)))..))))))...(((((((...))))))).))))...........))))............ ( -22.70) >DroYak_CAF1 29494 116 + 1 UAGGAGGAAAUAUUUGUCAUAACAGAGGACA---AAUAACUGCCUGCAAGGAGG-CACUCAUUAGGCAGUCGGAAUUGCAGCAGUUCCACUUCAAUUUCAUUACUUCCACAUCUUCGAUU ..(((((...((((((((.........))))---))))((((((((....(((.-..)))..)))))))).(((((((...)))))))...............)))))............ ( -32.70) >consensus UAGGAGCAAAUAAAUGUCACAACAGAGGAAGAGCAAGAACUGCCUGCAAAGAGGGAACUCAUUAGGCAGUCGGAAUUGCAGCGGUUCCACUUCAAUUUCAUUACUUCCACAUCUUCGAUU ..((((........(((....)))..(((((.......((((((((....(((....)))..)))))))).(((((((.((.(....).)).)))))))....)))))....)))).... (-25.08 = -26.64 + 1.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:11:15 2006