
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,425,777 – 4,426,095 |
| Length | 318 |
| Max. P | 0.931669 |

| Location | 4,425,777 – 4,425,895 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.64 |
| Mean single sequence MFE | -31.36 |
| Consensus MFE | -26.08 |
| Energy contribution | -26.52 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.855312 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4425777 118 + 22224390 AGCUGUUUAUCCCCCACCCCAGACAAUCGGCAAAGUAAACAACGGCUUGAGCUC-GACACCC-AUCGCAAUUCAUGGACUAUACACGCAGCCGGAGUCAAGUGGGUCAAAAUCAGCAUAC .((((.......(((((....(((..(((((...........((((....)).)-)....((-((........))))............))))).)))..))))).......)))).... ( -29.94) >DroSec_CAF1 28155 119 + 1 AGCUGUUUAUCCCCCACCCCAGACGAUCGGCUAAGUAAACAACGGCUUGAGCUC-GACGCCCGAUCGCAAUUCAUGGACUACACACUCAGCCGGAGUCAAGUGGGUCAAAAUCAGCAUAC .((((.......(((((....(((..((((((.(((.......((((((....)-)).))).((.......))...........))).)))))).)))..))))).......)))).... ( -34.14) >DroSim_CAF1 29488 119 + 1 AGCUGUUUAUCCCCCACCCCAGACGAUCGGCUAAGUAAACAACGGCUUGAGCUC-GACGCCCGAUCGCAAUUCAUGGACUACACACUCAGCCGGAGUCAAGUGGGUCAAAAUCAGCAUAC .((((.......(((((....(((..((((((.(((.......((((((....)-)).))).((.......))...........))).)))))).)))..))))).......)))).... ( -34.14) >DroEre_CAF1 29145 120 + 1 AGCUGUUUAUUCCCCACCCCACACAAUCGGCAAAGUAAACAACGACUCGAGCUCCAACGCCCGAUCGCAAUUCAUGGACUACACACUCAGCCGGAGUCAAGUGGGUCAAAAUCAGCAUAC .((((.......(((((...((....(((((..(((......(((.(((.((......)).)))))).......((.....)).)))..))))).))...))))).......)))).... ( -29.04) >DroYak_CAF1 28752 119 + 1 AGCUGUUUAUCCCCCACCCCAGACACUCGGCAAAGUAAACAACGACUCGAGCUC-GACGCCCGAUCGCAAUUCAUGGACUACACACUCAGCCUGAGUCAAGUGGGUCAAAAUCAGCAUAC .((((.......(((((.......(((((((..(((......(((.(((.((..-...)).)))))).......((.....)).)))..))).))))...))))).......)))).... ( -29.54) >consensus AGCUGUUUAUCCCCCACCCCAGACAAUCGGCAAAGUAAACAACGGCUUGAGCUC_GACGCCCGAUCGCAAUUCAUGGACUACACACUCAGCCGGAGUCAAGUGGGUCAAAAUCAGCAUAC .((((.......(((((....(((..(((((..(((......(((.(((.((......)).)))))).......((.....)).)))..))))).)))..))))).......)))).... (-26.08 = -26.52 + 0.44)


| Location | 4,425,777 – 4,425,895 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.64 |
| Mean single sequence MFE | -41.78 |
| Consensus MFE | -35.06 |
| Energy contribution | -35.70 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.588929 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4425777 118 - 22224390 GUAUGCUGAUUUUGACCCACUUGACUCCGGCUGCGUGUAUAGUCCAUGAAUUGCGAU-GGGUGUC-GAGCUCAAGCCGUUGUUUACUUUGCCGAUUGUCUGGGGUGGGGGAUAAACAGCU ....((((.((.(..((((((.(((..((((..((((.......))))....(((((-((.((..-.....))..))))))).......))))...)))...))))))..).)).)))). ( -35.10) >DroSec_CAF1 28155 119 - 1 GUAUGCUGAUUUUGACCCACUUGACUCCGGCUGAGUGUGUAGUCCAUGAAUUGCGAUCGGGCGUC-GAGCUCAAGCCGUUGUUUACUUAGCCGAUCGUCUGGGGUGGGGGAUAAACAGCU ....((((.((.(..((((((.(((..((((((((((((((((......))))))...((((...-..))))...........))))))))))...)))...))))))..).)).)))). ( -42.50) >DroSim_CAF1 29488 119 - 1 GUAUGCUGAUUUUGACCCACUUGACUCCGGCUGAGUGUGUAGUCCAUGAAUUGCGAUCGGGCGUC-GAGCUCAAGCCGUUGUUUACUUAGCCGAUCGUCUGGGGUGGGGGAUAAACAGCU ....((((.((.(..((((((.(((..((((((((((((((((......))))))...((((...-..))))...........))))))))))...)))...))))))..).)).)))). ( -42.50) >DroEre_CAF1 29145 120 - 1 GUAUGCUGAUUUUGACCCACUUGACUCCGGCUGAGUGUGUAGUCCAUGAAUUGCGAUCGGGCGUUGGAGCUCGAGUCGUUGUUUACUUUGCCGAUUGUGUGGGGUGGGGAAUAAACAGCU ....((((.((.(..(((((((.((.(((((.(((((((.....)))((((.((((((((((......)))))).)))).)))))))).))))...).)).)))))))..).)).)))). ( -45.40) >DroYak_CAF1 28752 119 - 1 GUAUGCUGAUUUUGACCCACUUGACUCAGGCUGAGUGUGUAGUCCAUGAAUUGCGAUCGGGCGUC-GAGCUCGAGUCGUUGUUUACUUUGCCGAGUGUCUGGGGUGGGGGAUAAACAGCU ....((((.((.(..((((((.(((.(.(((.(((((((.....)))((((.((((((((((...-..)))))).)))).)))))))).)))..).)))...))))))..).)).)))). ( -43.40) >consensus GUAUGCUGAUUUUGACCCACUUGACUCCGGCUGAGUGUGUAGUCCAUGAAUUGCGAUCGGGCGUC_GAGCUCAAGCCGUUGUUUACUUUGCCGAUUGUCUGGGGUGGGGGAUAAACAGCU ....((((.((.(..((((((.(((..((((.(((((((((((......))))))...((((......))))...........))))).))))...)))...))))))..).)).)))). (-35.06 = -35.70 + 0.64)


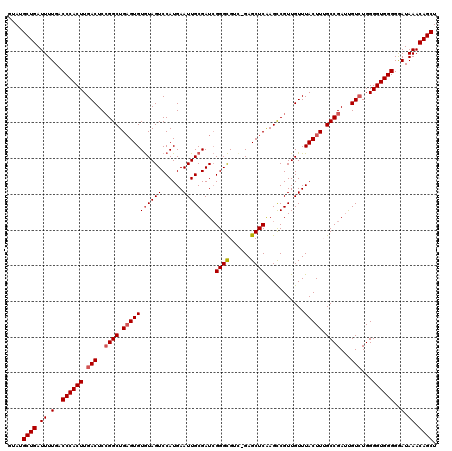
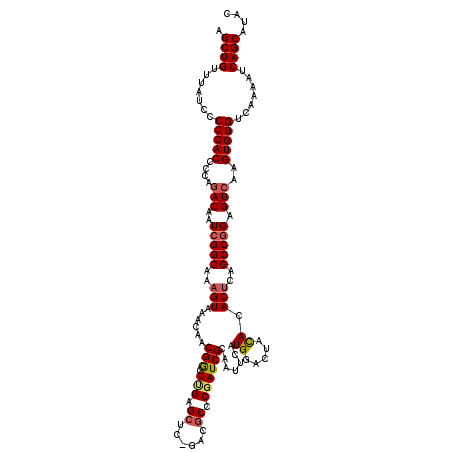
| Location | 4,425,895 – 4,426,015 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.12 |
| Mean single sequence MFE | -38.08 |
| Consensus MFE | -31.57 |
| Energy contribution | -32.65 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.910642 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4425895 120 + 22224390 AAAUCCGCAACUGAUGUCGCUGAUUUGAUCGCACUGGCGACGAUUCGUAUAUGGUAUAUGUUAUCGGAUCGGAUCGGAUCGGGGUAAUCCGAUUGGAUCACGCGAUGGGAUCAACUAAGU (((((.((.((....)).)).)))))((((.((.(.(((.(((((((.(((((...)))))...)))))))((((.(((((((....))))))).)))).))).))).))))........ ( -43.60) >DroSec_CAF1 28274 113 + 1 AAAUCCGCAACUGAUGUCGCUGAUUUGAUCGCACUGGCGACGAUUGGUAUAUGG-------UAUCGGAUCGGAACGGAUCGGGGGAAUCCGAUUGGAUCACGCGAUGGGAUCAACUAAGU (((((.((.((....)).)).)))))((((.((.(.(((.((((((((((...)-------)))).)))))((.(.(((((((....))))))).).)).))).))).))))........ ( -35.80) >DroSim_CAF1 29607 113 + 1 AAAUCCGCAACUGAUGUCGCUGAUUUGAUCGCACUGGCGGCGAUUGGUAUAUGG-------UAUCGGAUCGGAUCGGAUCGGGGGAAUCCGAUUGGAUCACGCGAUGGGAUCAACUAAGU (((((.((.((....)).)).)))))((((.((.(.(((.((((((((((...)-------)))).)))))((((.(((((((....))))))).)))).))).))).))))........ ( -40.30) >DroEre_CAF1 29265 110 + 1 AAAUCCGCAACUGAUGUCGCUGAUUUGAUCGCACUGGCGACGAUUGGUAUAUGGUAU-------C-GAUCGCAUAGUAUCGGGG--AUCCGAUUGGAUCACGCGAUGGGAUCAACUAAGU (((((.((.((....)).)).)))))((((.(((((..(.((((((((((...))))-------)-)))))).))))((((.((--((((....))))).).))))).))))........ ( -36.60) >DroYak_CAF1 28871 118 + 1 AAAUCCGCAACUGAUGUCGCUGAUUUGAUCGCACUGGCGACGAUUGGUAUAUGGUAUAUGGUAUCGGAUCGUAUUGUAUCGGGG--AUCCGAUUGGAUCACGCGAUGGGAUCAACUAAGU ......((.((....)).))((((((.(((((.((((..(((((((((...((((((...)))))).)))).))))).)))).(--((((....)))))..))))).))))))....... ( -34.10) >consensus AAAUCCGCAACUGAUGUCGCUGAUUUGAUCGCACUGGCGACGAUUGGUAUAUGGUAU____UAUCGGAUCGGAUCGGAUCGGGG_AAUCCGAUUGGAUCACGCGAUGGGAUCAACUAAGU (((((.((.((....)).)).)))))((((.((.(.(((.(((((.(.................).)))))((((.(((((((....))))))).)))).))).))).))))........ (-31.57 = -32.65 + 1.08)


| Location | 4,425,895 – 4,426,015 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.12 |
| Mean single sequence MFE | -30.96 |
| Consensus MFE | -24.40 |
| Energy contribution | -24.40 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.931669 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4425895 120 - 22224390 ACUUAGUUGAUCCCAUCGCGUGAUCCAAUCGGAUUACCCCGAUCCGAUCCGAUCCGAUAACAUAUACCAUAUACGAAUCGUCGCCAGUGCGAUCAAAUCAGCGACAUCAGUUGCGGAUUU ......((((((.(((.(((.((((..((((((((.....))))))))..))))((((....((((...))))...)))).)))..))).))))))(((.(((((....))))).))).. ( -32.30) >DroSec_CAF1 28274 113 - 1 ACUUAGUUGAUCCCAUCGCGUGAUCCAAUCGGAUUCCCCCGAUCCGUUCCGAUCCGAUA-------CCAUAUACCAAUCGUCGCCAGUGCGAUCAAAUCAGCGACAUCAGUUGCGGAUUU ......((((((.(((.(((.((((....((((((.....))))))....))))((((.-------..........)))).)))..))).))))))(((.(((((....))))).))).. ( -29.30) >DroSim_CAF1 29607 113 - 1 ACUUAGUUGAUCCCAUCGCGUGAUCCAAUCGGAUUCCCCCGAUCCGAUCCGAUCCGAUA-------CCAUAUACCAAUCGCCGCCAGUGCGAUCAAAUCAGCGACAUCAGUUGCGGAUUU ......((((((.(((.(((.((((..((((((((.....))))))))..))))((((.-------..........)))).)))..))).))))))(((.(((((....))))).))).. ( -32.60) >DroEre_CAF1 29265 110 - 1 ACUUAGUUGAUCCCAUCGCGUGAUCCAAUCGGAU--CCCCGAUACUAUGCGAUC-G-------AUACCAUAUACCAAUCGUCGCCAGUGCGAUCAAAUCAGCGACAUCAGUUGCGGAUUU ......((((((.(((((.(.(((((....))))--).)))))(((..((((.(-(-------((...........)))))))).)))).))))))(((.(((((....))))).))).. ( -31.80) >DroYak_CAF1 28871 118 - 1 ACUUAGUUGAUCCCAUCGCGUGAUCCAAUCGGAU--CCCCGAUACAAUACGAUCCGAUACCAUAUACCAUAUACCAAUCGUCGCCAGUGCGAUCAAAUCAGCGACAUCAGUUGCGGAUUU ......((((((.(((.(((((((...(((((((--(.............))))))))...((((...))))....)))).)))..))).))))))(((.(((((....))))).))).. ( -28.82) >consensus ACUUAGUUGAUCCCAUCGCGUGAUCCAAUCGGAUU_CCCCGAUCCGAUCCGAUCCGAUA____AUACCAUAUACCAAUCGUCGCCAGUGCGAUCAAAUCAGCGACAUCAGUUGCGGAUUU ......((((((.(((.(((((((...(((((......))))).(.....).........................)))).)))..))).))))))(((.(((((....))))).))).. (-24.40 = -24.40 + 0.00)


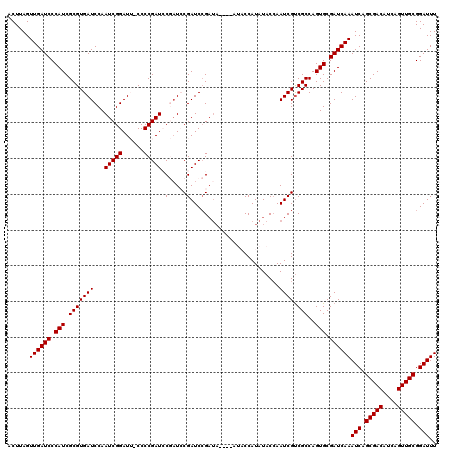
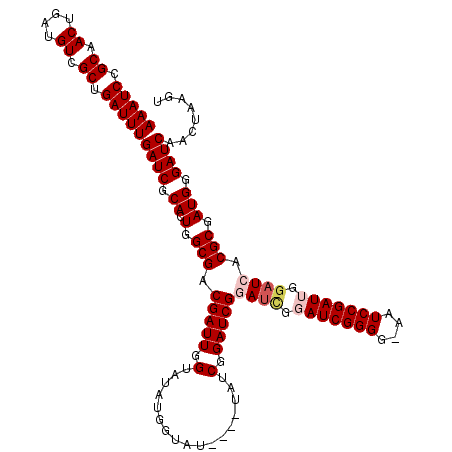
| Location | 4,425,935 – 4,426,055 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.12 |
| Mean single sequence MFE | -24.28 |
| Consensus MFE | -18.14 |
| Energy contribution | -19.14 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.860238 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4425935 120 - 22224390 UCAAUAUUCAACGCAUUACAAUUGCAAUAAGUUCCAGGCAACUUAGUUGAUCCCAUCGCGUGAUCCAAUCGGAUUACCCCGAUCCGAUCCGAUCCGAUAACAUAUACCAUAUACGAAUCG ((.....((((((((.......)))..((((((......)))))))))))........((.((((..((((((((.....))))))))..))))))..................)).... ( -26.70) >DroSec_CAF1 28314 113 - 1 UCAAUAUUCAACGCAUUACAAUUGCAAUAAGUUCCAGGCAACUUAGUUGAUCCCAUCGCGUGAUCCAAUCGGAUUCCCCCGAUCCGUUCCGAUCCGAUA-------CCAUAUACCAAUCG .......((((((((.......)))..((((((......)))))))))))........((.((((....((((((.....))))))....))))))...-------.............. ( -23.10) >DroSim_CAF1 29647 113 - 1 UCAAUAUUCAACGCAUUACAAUUGCAAUAAGUUCCAGGCAACUUAGUUGAUCCCAUCGCGUGAUCCAAUCGGAUUCCCCCGAUCCGAUCCGAUCCGAUA-------CCAUAUACCAAUCG .......((((((((.......)))..((((((......)))))))))))........((.((((..((((((((.....))))))))..))))))...-------.............. ( -26.40) >DroEre_CAF1 29305 110 - 1 UGAAUAUUCAACGCAUUACAAUUGCAAUAAGUUCCAGGCAACUUAGUUGAUCCCAUCGCGUGAUCCAAUCGGAU--CCCCGAUACUAUGCGAUC-G-------AUACCAUAUACCAAUCG .((....((((((((.......)))..((((((......)))))))))))..(.((((((((.....(((((..--..)))))..)))))))).-)-------..............)). ( -24.10) >DroYak_CAF1 28911 118 - 1 UCAAUAUUCAACGCAUUACAAUUGCAAUAAGUUCCAGGCAACUUAGUUGAUCCCAUCGCGUGAUCCAAUCGGAU--CCCCGAUACAAUACGAUCCGAUACCAUAUACCAUAUACCAAUCG .......((((((((.......)))..((((((......)))))))))))....((((.(.(((((....))))--).)))))..................................... ( -21.10) >consensus UCAAUAUUCAACGCAUUACAAUUGCAAUAAGUUCCAGGCAACUUAGUUGAUCCCAUCGCGUGAUCCAAUCGGAUU_CCCCGAUCCGAUCCGAUCCGAUA____AUACCAUAUACCAAUCG .......((((((((.......)))..((((((......)))))))))))....((((...((((..(((((......)))))..))))))))........................... (-18.14 = -19.14 + 1.00)



| Location | 4,425,975 – 4,426,095 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.00 |
| Mean single sequence MFE | -24.64 |
| Consensus MFE | -23.40 |
| Energy contribution | -23.40 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.841029 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4425975 120 - 22224390 CCGAACGAAUCUCAACCCCAAGAGCAUGCUGGCUCUCUCAUCAAUAUUCAACGCAUUACAAUUGCAAUAAGUUCCAGGCAACUUAGUUGAUCCCAUCGCGUGAUCCAAUCGGAUUACCCC .....(((............(((((......)))))...........((((((((.......)))..((((((......))))))))))).....))).(((((((....)))))))... ( -26.50) >DroSec_CAF1 28347 120 - 1 CCGAACGAAUCUCAACCCCAAGAGCAUGCUGGCUCUCUCAUCAAUAUUCAACGCAUUACAAUUGCAAUAAGUUCCAGGCAACUUAGUUGAUCCCAUCGCGUGAUCCAAUCGGAUUCCCCC ((((..(.(((.(.......(((((......)))))...........((((((((.......)))..((((((......))))))))))).........).))).)..))))........ ( -23.40) >DroSim_CAF1 29680 120 - 1 CCGAACGAAUCUCAACCCCAAGAGCAUGCUGGCUCUCUCAUCAAUAUUCAACGCAUUACAAUUGCAAUAAGUUCCAGGCAACUUAGUUGAUCCCAUCGCGUGAUCCAAUCGGAUUCCCCC ((((..(.(((.(.......(((((......)))))...........((((((((.......)))..((((((......))))))))))).........).))).)..))))........ ( -23.40) >DroEre_CAF1 29337 118 - 1 CCGAACGAAUCUCAACCCCAAGAGCAGGCUGGCUCUCUCAUGAAUAUUCAACGCAUUACAAUUGCAAUAAGUUCCAGGCAACUUAGUUGAUCCCAUCGCGUGAUCCAAUCGGAU--CCCC ((((................(((((......))))).(((((.....((((((((.......)))..((((((......)))))))))))........))))).....))))..--.... ( -25.72) >DroYak_CAF1 28951 118 - 1 CCGAACGAAUCUCAACCCCAAGAGCAGGCUGGCUCUCUCAUCAAUAUUCAACGCAUUACAAUUGCAAUAAGUUCCAGGCAACUUAGUUGAUCCCAUCGCGUGAUCCAAUCGGAU--CCCC ....(((.............(((((......)))))...........((((((((.......)))..((((((......)))))))))))........)))(((((....))))--)... ( -24.20) >consensus CCGAACGAAUCUCAACCCCAAGAGCAUGCUGGCUCUCUCAUCAAUAUUCAACGCAUUACAAUUGCAAUAAGUUCCAGGCAACUUAGUUGAUCCCAUCGCGUGAUCCAAUCGGAUU_CCCC ((((..(.(((.(.......(((((......)))))...........((((((((.......)))..((((((......))))))))))).........).))).)..))))........ (-23.40 = -23.40 + 0.00)

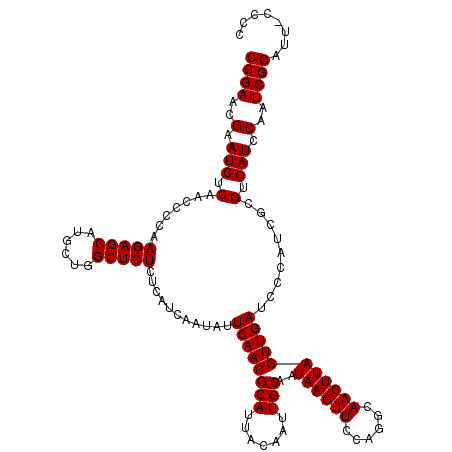

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:11:13 2006