
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,425,419 – 4,425,538 |
| Length | 119 |
| Max. P | 0.999958 |

| Location | 4,425,419 – 4,425,538 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.29 |
| Mean single sequence MFE | -34.48 |
| Consensus MFE | -31.80 |
| Energy contribution | -32.00 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.23 |
| Structure conservation index | 0.92 |
| SVM decision value | 4.87 |
| SVM RNA-class probability | 0.999958 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4425419 119 + 22224390 AUUAAACAUCUUGUGUGGAGCUUUAACGCCGUUUUACCCGGUAUCUGUAUCUCGUUCCGCUCGGUAUUUGUCUUUGUCGGCGUUUUGAUGUUU-UUAUCUCUGCCCGAUGCUCCACAUAC ...........((((((((((......((((....((..((((....))))..))......))))..........(((((.((...((((...-.))))...))))))))))))))))). ( -33.20) >DroSec_CAF1 27812 119 + 1 AUUAAACAUCUUGUGUGGAGCUUUAACGCCGUUUUACCCGGUAUCUGUAUCUCGUUCCGCCCGGUAUUUGUCUUUGUCGGCGUUUUGAUGUUU-UUAUCUCUGCCCGAUGCUCCACAUAC ...........((((((((((.............((((.(((................))).)))).........(((((.((...((((...-.))))...))))))))))))))))). ( -33.19) >DroSim_CAF1 29145 119 + 1 AUUAAACAUCUUGUGUGGAGCUUUAACGCCGUUUUACCCGGUAUCUGUAUCUCGUUCCGCCCGGUAUUUGUCUUUGUCGGCGUUUUGAUGUUU-UUAUCUCUGCCCGAUGCUCCACAUAC ...........((((((((((.............((((.(((................))).)))).........(((((.((...((((...-.))))...))))))))))))))))). ( -33.19) >DroEre_CAF1 28794 115 + 1 AUUAAACAUCUUGUGUGGAGCUUUAACGCCGUUUUACCCGGUAUCUGUAUCUUGU----CCCGGUGUUUGUCUUUGUCGGCGUCUUGAUGUUU-UUAUCUCUGCCCGAUGCUCCACAUAC ...........((((((((((...(((((((....((..((((....))))..))----..))))))).......(((((.((...((((...-.))))...))))))))))))))))). ( -35.50) >DroYak_CAF1 28424 116 + 1 AUUAUACAUCUUGUGUGGAGCUUUAACGCCGUUUUACCCGGUAUCUGUAUCUUGU----CCCGGUGUUUGUCUUUGUCGGCGGUUUGAUGUUUUUUAUCUCUGCCCGAUGCUCCACAUAC ...........((((((((((...(((((((....((..((((....))))..))----..))))))).......((((((((...((((.....)))).))).))))))))))))))). ( -37.30) >consensus AUUAAACAUCUUGUGUGGAGCUUUAACGCCGUUUUACCCGGUAUCUGUAUCUCGUUCCGCCCGGUAUUUGUCUUUGUCGGCGUUUUGAUGUUU_UUAUCUCUGCCCGAUGCUCCACAUAC ...........((((((((((......((((....((..((((....))))..))......))))..........(((((.((...((((.....))))...))))))))))))))))). (-31.80 = -32.00 + 0.20)

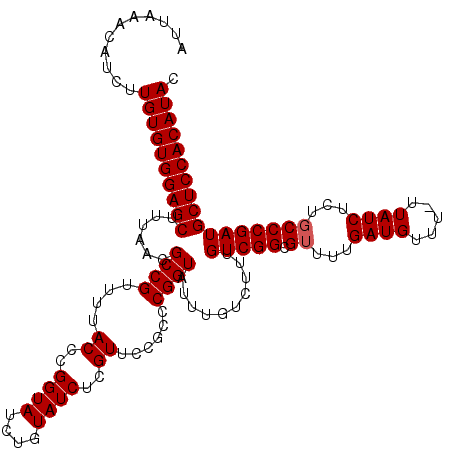
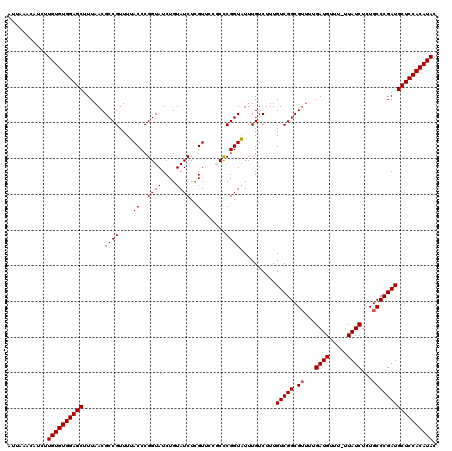
| Location | 4,425,419 – 4,425,538 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.29 |
| Mean single sequence MFE | -29.72 |
| Consensus MFE | -27.99 |
| Energy contribution | -28.23 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.94 |
| SVM decision value | 3.79 |
| SVM RNA-class probability | 0.999616 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4425419 119 - 22224390 GUAUGUGGAGCAUCGGGCAGAGAUAA-AAACAUCAAAACGCCGACAAAGACAAAUACCGAGCGGAACGAGAUACAGAUACCGGGUAAAACGGCGUUAAAGCUCCACACAAGAUGUUUAAU ...((((((((.(((..(...(((..-....)))....(((((..............)).))))..)))......((..(((.......)))..))...))))))))............. ( -28.54) >DroSec_CAF1 27812 119 - 1 GUAUGUGGAGCAUCGGGCAGAGAUAA-AAACAUCAAAACGCCGACAAAGACAAAUACCGGGCGGAACGAGAUACAGAUACCGGGUAAAACGGCGUUAAAGCUCCACACAAGAUGUUUAAU ...(((((((((((.......)))..-.........(((((((......((.....((((...................))))))....)))))))...))))))))............. ( -29.91) >DroSim_CAF1 29145 119 - 1 GUAUGUGGAGCAUCGGGCAGAGAUAA-AAACAUCAAAACGCCGACAAAGACAAAUACCGGGCGGAACGAGAUACAGAUACCGGGUAAAACGGCGUUAAAGCUCCACACAAGAUGUUUAAU ...(((((((((((.......)))..-.........(((((((......((.....((((...................))))))....)))))))...))))))))............. ( -29.91) >DroEre_CAF1 28794 115 - 1 GUAUGUGGAGCAUCGGGCAGAGAUAA-AAACAUCAAGACGCCGACAAAGACAAACACCGGG----ACAAGAUACAGAUACCGGGUAAAACGGCGUUAAAGCUCCACACAAGAUGUUUAAU ...(((((((((((.......)))..-.........(((((((.....(.....).((((.----..............))))......)))))))...))))))))............. ( -31.66) >DroYak_CAF1 28424 116 - 1 GUAUGUGGAGCAUCGGGCAGAGAUAAAAAACAUCAAACCGCCGACAAAGACAAACACCGGG----ACAAGAUACAGAUACCGGGUAAAACGGCGUUAAAGCUCCACACAAGAUGUAUAAU ...(((((((((((.......)))..............(((((.....(.....).((((.----..............))))......))))).....))))))))............. ( -28.56) >consensus GUAUGUGGAGCAUCGGGCAGAGAUAA_AAACAUCAAAACGCCGACAAAGACAAAUACCGGGCGGAACGAGAUACAGAUACCGGGUAAAACGGCGUUAAAGCUCCACACAAGAUGUUUAAU ...(((((((((((.......)))............(((((((......((.....((((...................))))))....)))))))...))))))))............. (-27.99 = -28.23 + 0.24)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:11:08 2006