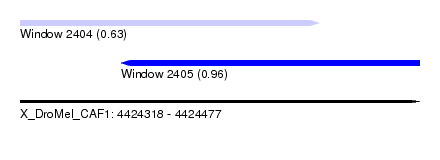
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,424,318 – 4,424,477 |
| Length | 159 |
| Max. P | 0.959536 |

| Location | 4,424,318 – 4,424,437 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.99 |
| Mean single sequence MFE | -32.88 |
| Consensus MFE | -29.50 |
| Energy contribution | -29.34 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.633965 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4424318 119 + 22224390 UCAAGCAAGUGCCCAUAUCAAAUGGAAUGCACACGGCCAAUGGGAGCAAUGAUGAUAUGUAUCUUUGGGCAAUUGGAGCACAACCGCCCACUGAUAACUGAGCUACCAGAAGGGGUAC-A ........((((((((((((........((.....))((.((....)).)).)))))).((((..(((((..(((.....)))..)))))..)))).(((......)))...))))))-. ( -32.70) >DroSec_CAF1 26712 120 + 1 UCAAGCAAGUGCCCAUAUCAAAUGGAAUGCACACGGCCAAUGGGAGCAAUGAUGAUAUGUAUCUUUGGGCAAUUGGAGCACAACCGCCCACUGAUAACUGAGCUACCAGAAGGGGUACCA ........((((((((((((........((.....))((.((....)).)).)))))).((((..(((((..(((.....)))..)))))..)))).(((......)))...)))))).. ( -31.80) >DroSim_CAF1 28050 120 + 1 UCAAGCAAGUGCCCAUAUCAAAUGGAAUGCACACGGCCAAUGGGAGCAAUGAUGAUAUGUAUCUUUGGGCAAUUGGAGCACAACCGCCCACUGAUAACCGAGCUACCAGAAGGGGUACCA ...(((..((((((((.....))))...)))).(((.((.((....)).))........((((..(((((..(((.....)))..)))))..)))).))).))).......((....)). ( -32.10) >DroEre_CAF1 27723 119 + 1 UCAAGCAAGUGCCCAUAUCAAGUGGAAUGCACACGGCCAAUGGGAGCAAUGAUGAUAUGUAUCUUUGGGCAAUUGGAGCACAACCGCCCACUGAUAACUGAGUAACGAGAAGGGGUAC-A ........((((((((((((.(((.......)))...((.((....)).)).)))))).((((..(((((..(((.....)))..)))))..))))................))))))-. ( -33.90) >DroYak_CAF1 27311 119 + 1 UCAAGCAAGUGCCUAUAUCAAGUGGAAUGCACACCGCCAAUGGGAGCAAUGAUGAUAUGUAUCUUUGGGCAAUUGGAGCACAACCGCCCACUGAUAACUGACUAACCAGAAAGGGUAC-A ........((((((((((((.((((........))))((.((....)).)).)))))).((((..(((((..(((.....)))..)))))..)))).(((......)))...))))))-. ( -33.90) >consensus UCAAGCAAGUGCCCAUAUCAAAUGGAAUGCACACGGCCAAUGGGAGCAAUGAUGAUAUGUAUCUUUGGGCAAUUGGAGCACAACCGCCCACUGAUAACUGAGCUACCAGAAGGGGUAC_A ........((((((((((((........((.....))((.((....)).)).)))))).((((..(((((..(((.....)))..)))))..))))................)))))).. (-29.50 = -29.34 + -0.16)



| Location | 4,424,358 – 4,424,477 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.32 |
| Mean single sequence MFE | -36.02 |
| Consensus MFE | -31.20 |
| Energy contribution | -31.12 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.959536 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4424358 119 - 22224390 AUUAGCAUGCAGUGCGUCUCAAAGGUAAAACUUUGGAAGGU-GUACCCCUUCUGGUAGCUCAGUUAUCAGUGGGCGGUUGUGCUCCAAUUGCCCAAAGAUACAUAUCAUCAUUGCUCCCA ........((((((..((.(((((......)))))))..((-(((......((((((((...))))))))((((((((((.....))))))))))....))))).....))))))..... ( -36.20) >DroSec_CAF1 26752 120 - 1 AUUAGCAUGCAGUGCGUCUCAAAGGUAAAACUUUGGAAGGUGGUACCCCUUCUGGUAGCUCAGUUAUCAGUGGGCGGUUGUGCUCCAAUUGCCCAAAGAUACAUAUCAUCAUUGCUCCCA ........(((((...((.(((((......))))))).(((((((......((((((((...))))))))((((((((((.....))))))))))........))))))))))))..... ( -37.60) >DroSim_CAF1 28090 120 - 1 AUUAGCAUGCAGUGCGUCUCAAAGGUAAAACUUUGGAAGGUGGUACCCCUUCUGGUAGCUCGGUUAUCAGUGGGCGGUUGUGCUCCAAUUGCCCAAAGAUACAUAUCAUCAUUGCUCCCA ........((((((.....(((((......)))))(((((.(....))))))(((((.....((.(((..((((((((((.....))))))))))..))))).))))).))))))..... ( -37.60) >DroEre_CAF1 27763 119 - 1 AUUAGCAUGCAGUGCGUCUCAAUGGUAAAACUUUGGAAGGU-GUACCCCUUCUCGUUACUCAGUUAUCAGUGGGCGGUUGUGCUCCAAUUGCCCAAAGAUACAUAUCAUCAUUGCUCCCA ........((((((.......((((((.(((...((((((.-.....)))))).))).....((.(((..((((((((((.....))))))))))..))))).))))))))))))..... ( -33.81) >DroYak_CAF1 27351 119 - 1 AUUAGCAUGCAGUGCGUCUCAAUGGUAAAACUUCGGAAGGU-GUACCCUUUCUGGUUAGUCAGUUAUCAGUGGGCGGUUGUGCUCCAAUUGCCCAAAGAUACAUAUCAUCAUUGCUCCCA ........((((((.......((((((.((((..((((((.-....)))))).)))).....((.(((..((((((((((.....))))))))))..))))).))))))))))))..... ( -34.91) >consensus AUUAGCAUGCAGUGCGUCUCAAAGGUAAAACUUUGGAAGGU_GUACCCCUUCUGGUAGCUCAGUUAUCAGUGGGCGGUUGUGCUCCAAUUGCCCAAAGAUACAUAUCAUCAUUGCUCCCA ........((((((..................((((((((.......)))))))).......((.(((..((((((((((.....))))))))))..))))).......))))))..... (-31.20 = -31.12 + -0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:11:05 2006