
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,423,988 – 4,424,113 |
| Length | 125 |
| Max. P | 0.989707 |

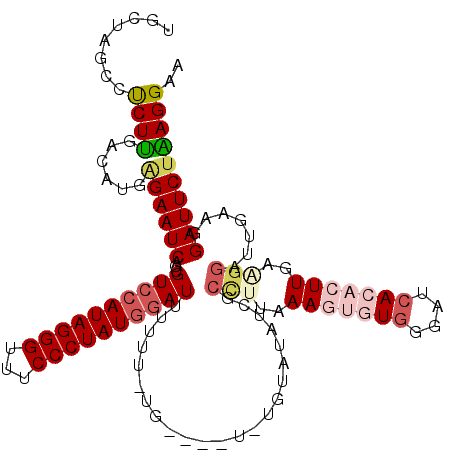
| Location | 4,423,988 – 4,424,103 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.98 |
| Mean single sequence MFE | -32.06 |
| Consensus MFE | -21.38 |
| Energy contribution | -23.26 |
| Covariance contribution | 1.88 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.577311 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4423988 115 + 22224390 UGCUAGCCCCUUGA-----GAAUCAGCGUCCAUAGGGUUUCCCUAUGGAUUUUUUUUGUAUAUAUGUAUAUCGCUUUGAAGUGUGGGAUCACACUUGAAGGAUUGAAGGAUUCUAAGGAA ........((((.(-----(((((...((((((((((...))))))))))....................((.((((.(((((((....))))))).))))...))..)))))))))).. ( -38.30) >DroSec_CAF1 26393 106 + 1 -GCUAGCCUCUUGACAUGGGAAUCAGCGUCCAUAGGGUUUCCCUAUGGAUUUUUU-UG----UUUGUAU--------GAAGUGUGGGAUCACACUUGAAGGAUUGAAGGAUUCUAAGGAA -.......((((......((((((...((((((((((...)))))))))).((((-.(----(((...(--------.(((((((....))))))).).)))).)))))))))).)))). ( -31.50) >DroSim_CAF1 27719 115 + 1 UGCUAGCCUCUUGACAUGGGAAUCAGCGUCCAUAGGGUUUCCCUAUGCAUUUUUU-UG----UAUGUAUAUCGCCUUAAAGUGUGGGAUCACACUUGAAGGAUUGAAGGAUUCUAAGGAA .(((..(((........)))....))).(((.((((((((...(((((((.....-..----.)))))))((.((((.(((((((....))))))).))))...)).)))))))).))). ( -31.10) >DroEre_CAF1 27351 108 + 1 UGCUAGCCUCUUGACAUGAGAAUCAGCGUCCAUAGGGUUUCCCUAUGGAUUUUUU-CG--------UAUACCCCCUUAAAGUGUGGGAUAA---UUGUCGGAUUGCAGGAUUCUGAGGAA ......((((..((((((((((.....((((((((((...)))))))))).))))-))--------)...(((.(.......).)))....---..)))(((((....))))).)))).. ( -32.70) >DroYak_CAF1 26962 108 + 1 UGCUAGUCUCUCGACAUGAGAAUCAGCGUCCAUAGGGUUUCCCUAUAGAUUUUUU-CG--------UAUAUCGCCUUCAAGUGUGGGAUCA---UUGUGGGAUUAAAGGAUUCUGAGGAA ......((((.......)))).((((.((((((((((...))))))...(((..(-(.--------((((...(((........)))....---.)))).))..))))))).)))).... ( -26.70) >consensus UGCUAGCCUCUUGACAUGAGAAUCAGCGUCCAUAGGGUUUCCCUAUGGAUUUUUU_UG____U_UGUAUAUCGCCUUAAAGUGUGGGAUCACACUUGAAGGAUUGAAGGAUUCUAAGGAA ........((((......((((((...((((((((((...)))))))))).......................(((..(((((((....)))))))..))).......)))))))))).. (-21.38 = -23.26 + 1.88)

| Location | 4,423,988 – 4,424,103 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.98 |
| Mean single sequence MFE | -28.90 |
| Consensus MFE | -18.68 |
| Energy contribution | -20.16 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.22 |
| Structure conservation index | 0.65 |
| SVM decision value | 2.18 |
| SVM RNA-class probability | 0.989707 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4423988 115 - 22224390 UUCCUUAGAAUCCUUCAAUCCUUCAAGUGUGAUCCCACACUUCAAAGCGAUAUACAUAUAUACAAAAAAAAUCCAUAGGGAAACCCUAUGGACGCUGAUUC-----UCAAGGGGCUAGCA .(((((((((((............(((((((....)))))))...((((((((....))))..........(((((((((...))))))))))))))))))-----).)))))....... ( -36.20) >DroSec_CAF1 26393 106 - 1 UUCCUUAGAAUCCUUCAAUCCUUCAAGUGUGAUCCCACACUUC--------AUACAAA----CA-AAAAAAUCCAUAGGGAAACCCUAUGGACGCUGAUUCCCAUGUCAAGAGGCUAGC- .......(((((............(((((((....))))))).--------.......----..-......(((((((((...)))))))))....)))))....(((....)))....- ( -26.00) >DroSim_CAF1 27719 115 - 1 UUCCUUAGAAUCCUUCAAUCCUUCAAGUGUGAUCCCACACUUUAAGGCGAUAUACAUA----CA-AAAAAAUGCAUAGGGAAACCCUAUGGACGCUGAUUCCCAUGUCAAGAGGCUAGCA ....((((..((.......((((.(((((((....))))))).))))...........----..-........(((((((...)))))))((((.((.....))))))..))..)))).. ( -27.90) >DroEre_CAF1 27351 108 - 1 UUCCUCAGAAUCCUGCAAUCCGACAA---UUAUCCCACACUUUAAGGGGGUAUA--------CG-AAAAAAUCCAUAGGGAAACCCUAUGGACGCUGAUUCUCAUGUCAAGAGGCUAGCA ..((((((((((..((..........---.((((((.(.......)))))))..--------..-......(((((((((...))))))))).)).))))))........))))...... ( -31.20) >DroYak_CAF1 26962 108 - 1 UUCCUCAGAAUCCUUUAAUCCCACAA---UGAUCCCACACUUGAAGGCGAUAUA--------CG-AAAAAAUCUAUAGGGAAACCCUAUGGACGCUGAUUCUCAUGUCGAGAGACUAGCA ......((((((.....(((((.(((---((......)).)))..)).)))...--------..-......(((((((((...)))))))))....))))))...(((....)))..... ( -23.20) >consensus UUCCUUAGAAUCCUUCAAUCCUUCAAGUGUGAUCCCACACUUCAAGGCGAUAUACA_A____CA_AAAAAAUCCAUAGGGAAACCCUAUGGACGCUGAUUCCCAUGUCAAGAGGCUAGCA .......(((((............(((((((....))))))).............................(((((((((...)))))))))....)))))....(((....)))..... (-18.68 = -20.16 + 1.48)

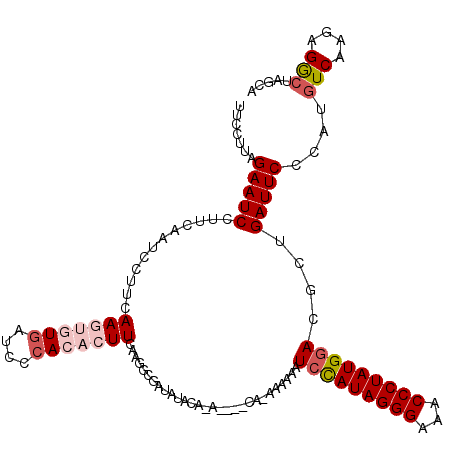

| Location | 4,424,023 – 4,424,113 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 86.79 |
| Mean single sequence MFE | -21.33 |
| Consensus MFE | -14.24 |
| Energy contribution | -15.80 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.67 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4424023 90 + 22224390 CCCUAUGGAUUUUUUUUGUAUAUAUGUAUAUCGCUUUGAAGUGUGGGAUCACACUUGAAGGAUUGAAGGAUUCUAAGGAAACUUUGUUUG .(((..(((((((((.(((((....)))))...((((.(((((((....))))))).))))...)))))))))..)))............ ( -21.10) >DroSec_CAF1 26432 77 + 1 CCCUAUGGAUUUUUU-UG----UUUGUAU--------GAAGUGUGGGAUCACACUUGAAGGAUUGAAGGAUUCUAAGGAAACUUUGAUUG .(((..(((((((((-.(----(((...(--------.(((((((....))))))).).)))).)))))))))..)))............ ( -19.20) >DroSim_CAF1 27759 85 + 1 CCCUAUGCAUUUUUU-UG----UAUGUAUAUCGCCUUAAAGUGUGGGAUCACACUUGAAGGAUUGAAGGAUUCUAAGGAAACUUUGAUUG .(((((((((.....-..----.)))))).((.((((.(((((((....))))))).))))...)))))..((.(((....))).))... ( -23.70) >consensus CCCUAUGGAUUUUUU_UG____UAUGUAUAUCGC_UUGAAGUGUGGGAUCACACUUGAAGGAUUGAAGGAUUCUAAGGAAACUUUGAUUG .(((..(((((((((..................((((.(((((((....))))))).))))...)))))))))..)))............ (-14.24 = -15.80 + 1.56)

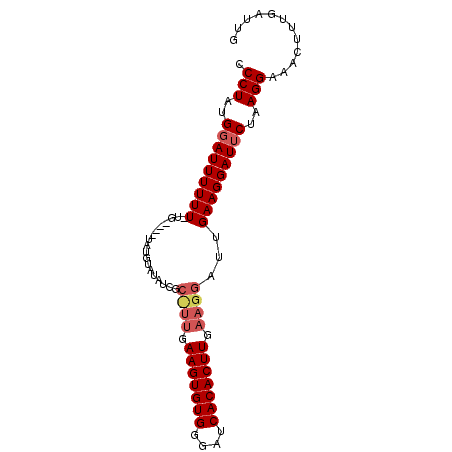
| Location | 4,424,023 – 4,424,113 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 86.79 |
| Mean single sequence MFE | -12.50 |
| Consensus MFE | -9.80 |
| Energy contribution | -9.80 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.665063 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4424023 90 - 22224390 CAAACAAAGUUUCCUUAGAAUCCUUCAAUCCUUCAAGUGUGAUCCCACACUUCAAAGCGAUAUACAUAUAUACAAAAAAAAUCCAUAGGG ............(((((((.....)).((((((.(((((((....)))))))..))).)))........................))))) ( -12.70) >DroSec_CAF1 26432 77 - 1 CAAUCAAAGUUUCCUUAGAAUCCUUCAAUCCUUCAAGUGUGAUCCCACACUUC--------AUACAAA----CA-AAAAAAUCCAUAGGG ............(((((((.....))........(((((((....))))))).--------.......----..-..........))))) ( -9.80) >DroSim_CAF1 27759 85 - 1 CAAUCAAAGUUUCCUUAGAAUCCUUCAAUCCUUCAAGUGUGAUCCCACACUUUAAGGCGAUAUACAUA----CA-AAAAAAUGCAUAGGG ............(((((((.....))...((((.(((((((....))))))).))))...........----..-..........))))) ( -15.00) >consensus CAAUCAAAGUUUCCUUAGAAUCCUUCAAUCCUUCAAGUGUGAUCCCACACUUCAA_GCGAUAUACAUA____CA_AAAAAAUCCAUAGGG ............(((((((.....))........(((((((....))))))).................................))))) ( -9.80 = -9.80 + -0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:11:03 2006