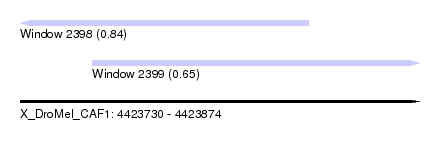
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,423,730 – 4,423,874 |
| Length | 144 |
| Max. P | 0.842564 |

| Location | 4,423,730 – 4,423,834 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.79 |
| Mean single sequence MFE | -35.88 |
| Consensus MFE | -27.50 |
| Energy contribution | -27.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.842564 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4423730 104 - 22224390 GAAACAAUUAGGCACCUGACUUCCUCAGGAUGCCAAAUAAACGGGAUGCAGACUGUAACCGAUCACGUUCCCACUGGG---GGAUGUGGGAUA--------CAGGGACAUGGGAU----- ..........(((((((((.....))))).))))...........((((...(((((.((...(((((((((....))---))))))))).))--------))).).))).....----- ( -35.50) >DroSec_CAF1 26133 113 - 1 GAAACAAUUAGGCACCUGACUUCCUCAGGAUGCCAAAUAAACGGGAUGCAGACUGUAACCGAUCACGUUCCCACUGGG--GGGAUGUGGGAUACAGGGAUACAGGGACAUGGGAU----- ..........(((((((((.....))))).))))...........((((...(((((.((..((((((((((.....)--)))))))))......))..))))).).))).....----- ( -36.70) >DroSim_CAF1 27459 113 - 1 GAAACAAUUAGGCACCUGACUUCCUCAGGAUGCCAAAUAAACGGGAUGCAGACUGUAACCGAUCACGUUCCCACUGGG--GGGAUGUGGGAUACAGGGAUACAGGGACAUGGGAU----- ..........(((((((((.....))))).))))...........((((...(((((.((..((((((((((.....)--)))))))))......))..))))).).))).....----- ( -36.70) >DroEre_CAF1 27081 117 - 1 GAAACAAUUAGGCACCUGACUUCCUCAGGAUGCCAAAUAAACGGGAUGCGGACUGUAACCGAUCACUUUCCCACUGG---GGGAUGUGGGACACAGGGAUACAGGGAUGUGGGAUGAACG ..........(((((((((.....))))).)))).......((...((((..(((((.((..((((.(((((....)---)))).))))......))..)))))...))))...)).... ( -34.50) >DroYak_CAF1 26718 101 - 1 GAAACAAUUAGGCACCUGACUUCCUCAGGAUGCCAAAUAAACGGGAUGCAGACUGUAACCGAUCACGUUCCCACCUGGGUGGGAUGUGGGAUACAGGAAUA------------------- ..........(((((((((.....))))).))))..................(((((.((...(((((.(((((....)))))))))))).))))).....------------------- ( -36.00) >consensus GAAACAAUUAGGCACCUGACUUCCUCAGGAUGCCAAAUAAACGGGAUGCAGACUGUAACCGAUCACGUUCCCACUGGG__GGGAUGUGGGAUACAGGGAUACAGGGACAUGGGAU_____ ((........(((((((((.....))))).)))).......(((..(((.....))).))).))....((((((...........))))))............................. (-27.50 = -27.50 + 0.00)


| Location | 4,423,756 – 4,423,874 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.43 |
| Mean single sequence MFE | -31.98 |
| Consensus MFE | -27.52 |
| Energy contribution | -27.56 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.653718 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4423756 118 + 22224390 --CCCAGUGGGAACGUGAUCGGUUACAGUCUGCAUCCCGUUUAUUUGGCAUCCUGAGGAAGUCAGGUGCCUAAUUGUUUCCGCCAUCAUCUGCAACCCCUGCUCCUAUUGAUUUCUUCCC --..((((((((..((((((((..(((((..((.....))......((((.(((((.....)))))))))..)))))..)))..)))))..(((.....))))))))))).......... ( -33.40) >DroSec_CAF1 26168 116 + 1 --CCCAGUGGGAACGUGAUCGGUUACAGUCUGCAUCCCGUUUAUUUGGCAUCCUGAGGAAGUCAGGUGCCUAAUUGUUUCCGCCAUCAUCCGCAACCCCUGCUCCUAUUGAUUUCUUC-- --..((((((((..((((((((..(((((..((.....))......((((.(((((.....)))))))))..)))))..)))..)))))..(((.....)))))))))))........-- ( -33.70) >DroSim_CAF1 27494 116 + 1 --CCCAGUGGGAACGUGAUCGGUUACAGUCUGCAUCCCGUUUAUUUGGCAUCCUGAGGAAGUCAGGUGCCUAAUUGUUUCCGCCAUCAUCUGCAACCCCUGCUCCUAUUGAUUUCUUC-- --..((((((((..((((((((..(((((..((.....))......((((.(((((.....)))))))))..)))))..)))..)))))..(((.....)))))))))))........-- ( -33.40) >DroEre_CAF1 27121 115 + 1 ---CCAGUGGGAAAGUGAUCGGUUACAGUCCGCAUCCCGUUUAUUUGGCAUCCUGAGGAAGUCAGGUGCCUAAUUGUUUCCGCCAUCAUCUCCAACCCCUGCU--UAUUGAUUUCUUCCC ---.(((.(((..(((((((((..(((((.((.....)).......((((.(((((.....)))))))))..)))))..)))..)))).))....))))))..--............... ( -28.40) >DroYak_CAF1 26739 118 + 1 ACCCAGGUGGGAACGUGAUCGGUUACAGUCUGCAUCCCGUUUAUUUGGCAUCCUGAGGAAGUCAGGUGCCUAAUUGUUUCCGCCAUCAUCUCCAACCCCUGCU--UAUUGAUUUCUUUCC .....((((((((((.((((((.......))).))).)........((((.(((((.....))))))))).....)))))))))...................--............... ( -31.00) >consensus __CCCAGUGGGAACGUGAUCGGUUACAGUCUGCAUCCCGUUUAUUUGGCAUCCUGAGGAAGUCAGGUGCCUAAUUGUUUCCGCCAUCAUCUGCAACCCCUGCUCCUAUUGAUUUCUUCCC ....(((.(((...((((((((..(((((..((.....))......((((.(((((.....)))))))))..)))))..)))..)))))......))))))................... (-27.52 = -27.56 + 0.04)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:11:00 2006