
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,423,437 – 4,423,674 |
| Length | 237 |
| Max. P | 0.999824 |

| Location | 4,423,437 – 4,423,554 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.97 |
| Mean single sequence MFE | -31.58 |
| Consensus MFE | -31.54 |
| Energy contribution | -31.54 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.49 |
| Structure conservation index | 1.00 |
| SVM decision value | 4.17 |
| SVM RNA-class probability | 0.999824 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4423437 117 + 22224390 ---AUGCAUUCACAAUUGCCCACUGGUGACAUAAUCCACUGGCAGCAGCCAACACAUUUUCUGUCACUGGAGACGCACUUGAACCUCCAGCAACAACAAUAUCCUGAAAGAGGUCAAGUG ---..(((........)))...(..((((((.(((....((((....))))....)))...))))))..).....(((((((.((((...((............))...))))))))))) ( -31.40) >DroSec_CAF1 25832 120 + 1 CAUAUGCAUUCACAAUUGCCCACUGGUGACAUAAUCCACUGGCAGCAGCCAACACAUUUUCUGUCACUGGAGACGCACUUGAACCUCCAGCAACAACAACAUCCUGAAAGAGGUCAAGUG .....(((........)))...(..((((((.(((....((((....))))....)))...))))))..).....(((((((.((((...((............))...))))))))))) ( -31.40) >DroSim_CAF1 27161 117 + 1 ---AUGCAUUCACAAUUGCCCACUGGUGACAUAAUCCACUGGCAGCAGCCAACACAUUUUCUGUCACUGGAGACGCACUUGAACCUCCAGCAACAACAACAUCCUGAAAGAGGUCAAGUG ---..(((........)))...(..((((((.(((....((((....))))....)))...))))))..).....(((((((.((((...((............))...))))))))))) ( -31.40) >DroEre_CAF1 26767 117 + 1 ---AUGCAUUCACAAUUGCCCACUGGUGACAUAAUCCACUGGCAGCAGCCAACACAUUUUCUGUCACUGGAGACGCACUUGAACCUCCAGCAACAACAACAUCCUGAAAGAGGUCAAGUG ---..(((........)))...(..((((((.(((....((((....))))....)))...))))))..).....(((((((.((((...((............))...))))))))))) ( -31.40) >DroYak_CAF1 26418 117 + 1 ---AUGCAUUCGCAAUUGCCCACUGGUGACAUAAUCCACUGGCAGCAGCCAACACAUUUUUUGUCACUGGAGACGCACUUGAACCUCCAGCAACAACAACAUCCUGAAAGAGGUCAAGUG ---.(((....)))........(..((((((.(((....((((....))))....)))...))))))..).....(((((((.((((...((............))...))))))))))) ( -32.30) >consensus ___AUGCAUUCACAAUUGCCCACUGGUGACAUAAUCCACUGGCAGCAGCCAACACAUUUUCUGUCACUGGAGACGCACUUGAACCUCCAGCAACAACAACAUCCUGAAAGAGGUCAAGUG .....(((........)))...(..((((((.(((....((((....))))....)))...))))))..).....(((((((.((((...((............))...))))))))))) (-31.54 = -31.54 + -0.00)



| Location | 4,423,437 – 4,423,554 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.97 |
| Mean single sequence MFE | -39.68 |
| Consensus MFE | -39.70 |
| Energy contribution | -39.54 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.92 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.90 |
| SVM RNA-class probability | 0.997630 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4423437 117 - 22224390 CACUUGACCUCUUUCAGGAUAUUGUUGUUGCUGGAGGUUCAAGUGCGUCUCCAGUGACAGAAAAUGUGUUGGCUGCUGCCAGUGGAUUAUGUCACCAGUGGGCAAUUGUGAAUGCAU--- ((((((((((((..(((.(......).)))..))))).))))))).(((..(.((((((...(((.(..((((....))))..).))).))))))..)..)))..............--- ( -39.50) >DroSec_CAF1 25832 120 - 1 CACUUGACCUCUUUCAGGAUGUUGUUGUUGCUGGAGGUUCAAGUGCGUCUCCAGUGACAGAAAAUGUGUUGGCUGCUGCCAGUGGAUUAUGUCACCAGUGGGCAAUUGUGAAUGCAUAUG ((((((((((((..(((.(......).)))..))))).))))))).(((..(.((((((...(((.(..((((....))))..).))).))))))..)..)))...((((....)))).. ( -40.00) >DroSim_CAF1 27161 117 - 1 CACUUGACCUCUUUCAGGAUGUUGUUGUUGCUGGAGGUUCAAGUGCGUCUCCAGUGACAGAAAAUGUGUUGGCUGCUGCCAGUGGAUUAUGUCACCAGUGGGCAAUUGUGAAUGCAU--- ((((((((((((..(((.(......).)))..))))).))))))).(((..(.((((((...(((.(..((((....))))..).))).))))))..)..)))..............--- ( -39.50) >DroEre_CAF1 26767 117 - 1 CACUUGACCUCUUUCAGGAUGUUGUUGUUGCUGGAGGUUCAAGUGCGUCUCCAGUGACAGAAAAUGUGUUGGCUGCUGCCAGUGGAUUAUGUCACCAGUGGGCAAUUGUGAAUGCAU--- ((((((((((((..(((.(......).)))..))))).))))))).(((..(.((((((...(((.(..((((....))))..).))).))))))..)..)))..............--- ( -39.50) >DroYak_CAF1 26418 117 - 1 CACUUGACCUCUUUCAGGAUGUUGUUGUUGCUGGAGGUUCAAGUGCGUCUCCAGUGACAAAAAAUGUGUUGGCUGCUGCCAGUGGAUUAUGUCACCAGUGGGCAAUUGCGAAUGCAU--- ..(((((......)))))......((((..(((((((..(....)..)))))))..))))...(((((((.((...(((((.(((.........))).).))))...)).)))))))--- ( -39.90) >consensus CACUUGACCUCUUUCAGGAUGUUGUUGUUGCUGGAGGUUCAAGUGCGUCUCCAGUGACAGAAAAUGUGUUGGCUGCUGCCAGUGGAUUAUGUCACCAGUGGGCAAUUGUGAAUGCAU___ ((((((((((((..(((.(......).)))..))))).))))))).(((..(.((((((...(((.(..((((....))))..).))).))))))..)..)))...(((....))).... (-39.70 = -39.54 + -0.16)


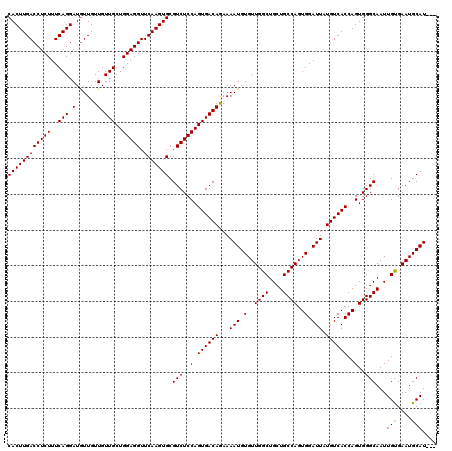
| Location | 4,423,474 – 4,423,594 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.97 |
| Mean single sequence MFE | -32.84 |
| Consensus MFE | -29.52 |
| Energy contribution | -30.86 |
| Covariance contribution | 1.34 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.760162 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4423474 120 + 22224390 GGCAGCAGCCAACACAUUUUCUGUCACUGGAGACGCACUUGAACCUCCAGCAACAACAAUAUCCUGAAAGAGGUCAAGUGAUUGGCCAUCUCAUUGUGUUUUCUCAUCAGGUGCGCACCU (((....)))((((((.....(((..((((((...(....)...))))))..))).............(((((((((....)))))).)))...))))))........((((....)))) ( -33.00) >DroSec_CAF1 25872 117 + 1 GGCAGCAGCCAACACAUUUUCUGUCACUGGAGACGCACUUGAACCUCCAGCAACAACAACAUCCUGAAAGAGGUCAAGUGAUUGGCCAUCUCAUUGUGUUUUCUCAUCAGGUGCGC---U (((.(((.((...........(((..((((((...(....)...))))))..)))..(((((..(((..((((((((....)))))).)))))..))))).........)))))))---) ( -32.20) >DroSim_CAF1 27198 117 + 1 GGCAGCAGCCAACACAUUUUCUGUCACUGGAGACGCACUUGAACCUCCAGCAACAACAACAUCCUGAAAGAGGUCAAGUGAUUGGCCAUCUCAUUGUGUUUUCUCAUCAGGUGCGC---U (((.(((.((...........(((..((((((...(....)...))))))..)))..(((((..(((..((((((((....)))))).)))))..))))).........)))))))---) ( -32.20) >DroEre_CAF1 26804 117 + 1 GGCAGCAGCCAACACAUUUUCUGUCACUGGAGACGCACUUGAACCUCCAGCAACAACAACAUCCUGAAAGAGGUCAAGUGAUUGGCCAUCUCAUUGUGUUUUCUCAUCAGGUGCGC---U (((.(((.((...........(((..((((((...(....)...))))))..)))..(((((..(((..((((((((....)))))).)))))..))))).........)))))))---) ( -32.20) >DroYak_CAF1 26455 117 + 1 GGCAGCAGCCAACACAUUUUUUGUCACUGGAGACGCACUUGAACCUCCAGCAACAACAACAUCCUGAAAGAGGUCAAGUGAUUGGCCAUCUCAUUGUGUUUUCUCAUCAGGUGCGC---C (((.(((.((..........((((..((((((...(....)...))))))..)))).(((((..(((..((((((((....)))))).)))))..))))).........)))))))---) ( -34.60) >consensus GGCAGCAGCCAACACAUUUUCUGUCACUGGAGACGCACUUGAACCUCCAGCAACAACAACAUCCUGAAAGAGGUCAAGUGAUUGGCCAUCUCAUUGUGUUUUCUCAUCAGGUGCGC___U (((....))).......(((((......)))))(((((((((......((((.(((((......))..(((((((((....)))))).)))..)))))))......)))))))))..... (-29.52 = -30.86 + 1.34)

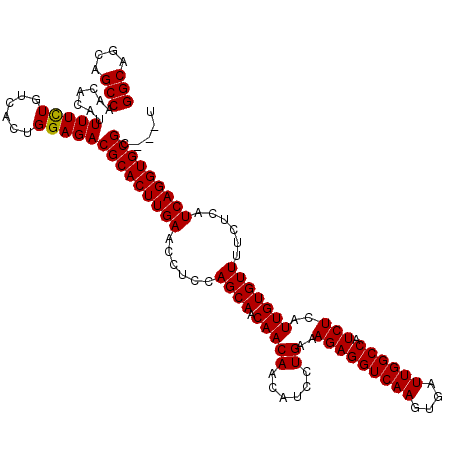
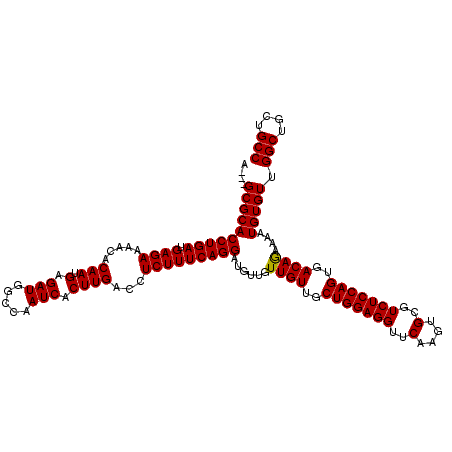
| Location | 4,423,474 – 4,423,594 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.97 |
| Mean single sequence MFE | -37.08 |
| Consensus MFE | -35.96 |
| Energy contribution | -35.80 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.677919 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4423474 120 - 22224390 AGGUGCGCACCUGAUGAGAAAACACAAUGAGAUGGCCAAUCACUUGACCUCUUUCAGGAUAUUGUUGUUGCUGGAGGUUCAAGUGCGUCUCCAGUGACAGAAAAUGUGUUGGCUGCUGCC .((((.((..((.((...........)).))..(((((((..(((((......)))))(((((.((((..(((((((..(....)..)))))))..))))..)))))))))))))))))) ( -36.50) >DroSec_CAF1 25872 117 - 1 A---GCGCACCUGAUGAGAAAACACAAUGAGAUGGCCAAUCACUUGACCUCUUUCAGGAUGUUGUUGUUGCUGGAGGUUCAAGUGCGUCUCCAGUGACAGAAAAUGUGUUGGCUGCUGCC (---((((((((((.((((.....(((.(.(((.....))).))))...)))))))))......((((..(((((((..(....)..)))))))..))))....))))))(((....))) ( -36.90) >DroSim_CAF1 27198 117 - 1 A---GCGCACCUGAUGAGAAAACACAAUGAGAUGGCCAAUCACUUGACCUCUUUCAGGAUGUUGUUGUUGCUGGAGGUUCAAGUGCGUCUCCAGUGACAGAAAAUGUGUUGGCUGCUGCC (---((((((((((.((((.....(((.(.(((.....))).))))...)))))))))......((((..(((((((..(....)..)))))))..))))....))))))(((....))) ( -36.90) >DroEre_CAF1 26804 117 - 1 A---GCGCACCUGAUGAGAAAACACAAUGAGAUGGCCAAUCACUUGACCUCUUUCAGGAUGUUGUUGUUGCUGGAGGUUCAAGUGCGUCUCCAGUGACAGAAAAUGUGUUGGCUGCUGCC (---((((((((((.((((.....(((.(.(((.....))).))))...)))))))))......((((..(((((((..(....)..)))))))..))))....))))))(((....))) ( -36.90) >DroYak_CAF1 26455 117 - 1 G---GCGCACCUGAUGAGAAAACACAAUGAGAUGGCCAAUCACUUGACCUCUUUCAGGAUGUUGUUGUUGCUGGAGGUUCAAGUGCGUCUCCAGUGACAAAAAAUGUGUUGGCUGCUGCC (---((((..((.((...........)).))..((((((.(((..(((.(((....))).))).((((..(((((((..(....)..)))))))..)))).....))))))))))).))) ( -38.20) >consensus A___GCGCACCUGAUGAGAAAACACAAUGAGAUGGCCAAUCACUUGACCUCUUUCAGGAUGUUGUUGUUGCUGGAGGUUCAAGUGCGUCUCCAGUGACAGAAAAUGUGUUGGCUGCUGCC ....((((((((((.((((.....(((.(.(((.....))).))))...)))))))))......((((..(((((((..(....)..)))))))..))))....))))).(((....))) (-35.96 = -35.80 + -0.16)

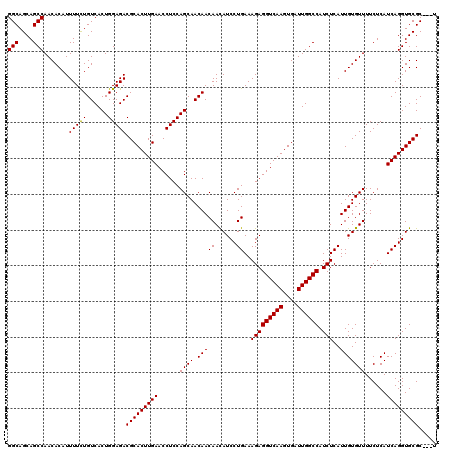
| Location | 4,423,554 – 4,423,674 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.12 |
| Mean single sequence MFE | -33.56 |
| Consensus MFE | -33.10 |
| Energy contribution | -33.34 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.662094 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4423554 120 + 22224390 AUUGGCCAUCUCAUUGUGUUUUCUCAUCAGGUGCGCACCUGAGAGGGCCUAAAGUCCAAAAACUCCGAGUGCCAAGUGGUUGGGCCCAAAGAAAUAUGAUACGCAACAUUGUGUAUGCAU ...((((...(((((.......(((.((((((....)))))))))(((((..(((......)))...)).))).)))))...)))).........((((((((((....))))))).))) ( -35.90) >DroSec_CAF1 25952 117 + 1 AUUGGCCAUCUCAUUGUGUUUUCUCAUCAGGUGCGC---UCAGAGGGCCUAAAGCCCAAAAACUCCGAGUGCCAAGUGGUUGGGCCCAAAGAAAUAUGAUACGCAACAUUGUGUAUGCAU ...((((...((((((((......)))...(.((((---((.((((((.....))))......)).))))))).)))))...)))).........((((((((((....))))))).))) ( -32.40) >DroSim_CAF1 27278 117 + 1 AUUGGCCAUCUCAUUGUGUUUUCUCAUCAGGUGCGC---UGAGAGGGCCUAAAGUCCAAAAACUCCGAGUGCCAAGUGGUUGGGCCCAAAGAAAUAUGAUACGCAACAUUGUGUAUGCAU ...((((.(((((.((..(((.......)))..)).---))))).))))....((((((..(((..(....)..)))..))))))..........((((((((((....))))))).))) ( -33.40) >DroEre_CAF1 26884 117 + 1 AUUGGCCAUCUCAUUGUGUUUUCUCAUCAGGUGCGC---UGAGAGGGCCUAAAGUCCAAAAACUCCGAGUGCAAAGUGGUUGGGCCCAAAGAAAUAUGAUACGCAACAUUGUGUAUGCAU ...((((.(((((.((..(((.......)))..)).---))))).))))....((((((..(((..(....)..)))..))))))..........((((((((((....))))))).))) ( -33.30) >DroYak_CAF1 26535 117 + 1 AUUGGCCAUCUCAUUGUGUUUUCUCAUCAGGUGCGC---CGAGAGGGCCUAAAGUCCAAAAACUCCGAGUGCAAAGUGGUUGGGCUCAAAGAAAUAUGAUACGCAACAUUGUGUAUGCAU ...((((.((((...((((..(((....))).))))---.)))).))))...(((((((..(((..(....)..)))..))))))).........((((((((((....))))))).))) ( -32.80) >consensus AUUGGCCAUCUCAUUGUGUUUUCUCAUCAGGUGCGC___UGAGAGGGCCUAAAGUCCAAAAACUCCGAGUGCCAAGUGGUUGGGCCCAAAGAAAUAUGAUACGCAACAUUGUGUAUGCAU ...((((.(((((..((((..(((....))).))))...))))).))))....((((((..(((..(....)..)))..))))))..........((((((((((....))))))).))) (-33.10 = -33.34 + 0.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:10:58 2006