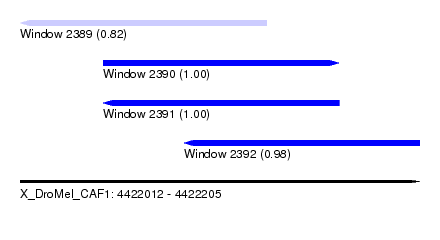
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,422,012 – 4,422,205 |
| Length | 193 |
| Max. P | 0.999647 |

| Location | 4,422,012 – 4,422,131 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.98 |
| Mean single sequence MFE | -28.19 |
| Consensus MFE | -25.53 |
| Energy contribution | -26.65 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.816043 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4422012 119 - 22224390 CUGGUCGUAAAGCCCAUGGCAUUGAUCAUAAUCGAUGCCACAGGAUAAUAAACUCUAGGAAUGCACUGCAA-GUCCAUUGAUGACUGCAGCAGGUUCAUAAAAAGAUACUGAAAGUAUCA ((((........((..((((((((((....))))))))))..))..........))))((((...(((((.-(((.......))))))))...)))).......(((((.....))))). ( -34.77) >DroSec_CAF1 24488 120 - 1 CUGGUCAUAAAGCCCAUGGCAUUGAUCAUAAUCGAUGCCACAGGAUAAUAAACUCUAGGAAUGCACUGCACUGUCCAUUGAUGACUGCAGCAGGUUCAUAAAAAGAUACUGAAAGAAUCA ..((((((....((..((((((((((....))))))))))..)).............(((.(((...)))...)))....))))))....(((..((.......))..)))......... ( -29.10) >DroSim_CAF1 25800 120 - 1 CUGGUCGUAAAGCCCAUGGCAUUGAUCAUAAUCGAUGCCACAGGAUAAUAAACUCUAGGAAUGCACUGCACUGUCCAUUGAUGACUGCAGCAGGUUCAUAAAAAGAUACUGAAAGAAUCA ..((((((....((..((((((((((....))))))))))..)).............(((.(((...)))...)))....))))))....(((..((.......))..)))......... ( -28.70) >DroEre_CAF1 25475 119 - 1 CUGGUCGUAAAGCCCAUGGCAUUGAUCAUAAUCGAAGGCGCAGGAUAAUAAACUCUAGGAAUGCACUGCAA-GUCCAUUGAAGGCUGCAGCAGGUUCAUAAAAAGAUUGGGAAAGAAUCG (..(((.......((.((((.(((((....)))))..)).))))..............((((...(((((.-(((.......))))))))...)))).......)))..).......... ( -23.10) >DroYak_CAF1 25056 119 - 1 CUGGUCGUAAAGCCCAUGGCAUUGAUCAUAAUCGAAGCCACAGGAUAAUAAACUCUAGGAAUGCACUGCAA-GUCCAUUGAAGACUGCAUCAGGUUCAUAAAAAGAUUCUGAAAAAAUCG ((((........((..((((.(((((....))))).))))..))..........))))((......((((.-(((.......)))))))((((..((.......))..)))).....)). ( -25.27) >consensus CUGGUCGUAAAGCCCAUGGCAUUGAUCAUAAUCGAUGCCACAGGAUAAUAAACUCUAGGAAUGCACUGCAA_GUCCAUUGAUGACUGCAGCAGGUUCAUAAAAAGAUACUGAAAGAAUCA ((((........((..((((((((((....))))))))))..))..........))))((((...(((((..(((.......))))))))...)))).......(((((.....))))). (-25.53 = -26.65 + 1.12)



| Location | 4,422,052 – 4,422,166 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.03 |
| Mean single sequence MFE | -38.86 |
| Consensus MFE | -33.72 |
| Energy contribution | -34.40 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.14 |
| Structure conservation index | 0.87 |
| SVM decision value | 3.83 |
| SVM RNA-class probability | 0.999647 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4422052 114 + 22224390 CAAUGGAC-UUGCAGUGCAUUCCUAGAGUUUAUUAUCCUGUGGCAUCGAUUAUGAUCAAUGCCAUGGGCUUUACGACCAGCAACUUGGAUCGUUAACUGGGAGUCCAUUAAACCG----- .(((((((-((.((((....................((((((((((.(((....))).))))))))))....((((((((....)))).))))..)))).)))))))))......----- ( -41.30) >DroSec_CAF1 24528 115 + 1 CAAUGGACAGUGCAGUGCAUUCCUAGAGUUUAUUAUCCUGUGGCAUCGAUUAUGAUCAAUGCCAUGGGCUUUAUGACCAGCAACUUGGAUCGUUAACUGGGAGUCCAUUAAACCG----- .(((((((.((((...))))((((((..........((((((((((.(((....))).))))))))))....((((((((....)))).))))...)))))))))))))......----- ( -39.60) >DroSim_CAF1 25840 115 + 1 CAAUGGACAGUGCAGUGCAUUCCUAGAGUUUAUUAUCCUGUGGCAUCGAUUAUGAUCAAUGCCAUGGGCUUUACGACCAGCAACUUGGAUCGUUAACUGGGAGUCCAUUAAACCG----- .(((((((.((((...))))((((((..........((((((((((.(((....))).))))))))))....((((((((....)))).))))...)))))))))))))......----- ( -41.40) >DroEre_CAF1 25515 114 + 1 CAAUGGAC-UUGCAGUGCAUUCCUAGAGUUUAUUAUCCUGCGCCUUCGAUUAUGAUCAAUGCCAUGGGCUUUACGACCAGCAACUUGGAUCGUUAACUGCGGGUCCAUUAAACCG----- .(((((((-(((((((((..(((..(((((......((((.((.((.(((....))))).)))).))(((........)))))))))))..))..))))))))))))))......----- ( -34.10) >DroYak_CAF1 25096 119 + 1 CAAUGGAC-UUGCAGUGCAUUCCUAGAGUUUAUUAUCCUGUGGCUUCGAUUAUGAUCAAUGCCAUGGGCUUUACGACCAGCAACUUGGAUCGUUAACUGGGAGUCCAUUAAACCGUAGCU .(((((((-((.((((....................((((((((...(((....)))...))))))))....((((((((....)))).))))..)))).)))))))))........... ( -37.90) >consensus CAAUGGAC_UUGCAGUGCAUUCCUAGAGUUUAUUAUCCUGUGGCAUCGAUUAUGAUCAAUGCCAUGGGCUUUACGACCAGCAACUUGGAUCGUUAACUGGGAGUCCAUUAAACCG_____ .(((((((..(((...))).((((((..........((((((((((.(((....))).))))))))))....((((((((....)))).))))...)))))))))))))........... (-33.72 = -34.40 + 0.68)


| Location | 4,422,052 – 4,422,166 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.03 |
| Mean single sequence MFE | -34.06 |
| Consensus MFE | -29.88 |
| Energy contribution | -30.32 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.74 |
| SVM RNA-class probability | 0.996739 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4422052 114 - 22224390 -----CGGUUUAAUGGACUCCCAGUUAACGAUCCAAGUUGCUGGUCGUAAAGCCCAUGGCAUUGAUCAUAAUCGAUGCCACAGGAUAAUAAACUCUAGGAAUGCACUGCAA-GUCCAUUG -----.....(((((((((..((((..((((.(((......)))))))....((..((((((((((....))))))))))..))....................))))..)-)))))))) ( -37.20) >DroSec_CAF1 24528 115 - 1 -----CGGUUUAAUGGACUCCCAGUUAACGAUCCAAGUUGCUGGUCAUAAAGCCCAUGGCAUUGAUCAUAAUCGAUGCCACAGGAUAAUAAACUCUAGGAAUGCACUGCACUGUCCAUUG -----.....(((((((((((.((((....((((.....(((........)))...((((((((((....))))))))))..))))....))))...))).(((...)))..)))))))) ( -34.00) >DroSim_CAF1 25840 115 - 1 -----CGGUUUAAUGGACUCCCAGUUAACGAUCCAAGUUGCUGGUCGUAAAGCCCAUGGCAUUGAUCAUAAUCGAUGCCACAGGAUAAUAAACUCUAGGAAUGCACUGCACUGUCCAUUG -----.....(((((((((((.((((.((((.(((......)))))))....((..((((((((((....))))))))))..))......))))...))).(((...)))..)))))))) ( -35.30) >DroEre_CAF1 25515 114 - 1 -----CGGUUUAAUGGACCCGCAGUUAACGAUCCAAGUUGCUGGUCGUAAAGCCCAUGGCAUUGAUCAUAAUCGAAGGCGCAGGAUAAUAAACUCUAGGAAUGCACUGCAA-GUCCAUUG -----.....((((((((..(((((...((.(((.((.(.((((((.....(((...))).(((((....))))).))).))).).........)).))).)).)))))..-)))))))) ( -29.50) >DroYak_CAF1 25096 119 - 1 AGCUACGGUUUAAUGGACUCCCAGUUAACGAUCCAAGUUGCUGGUCGUAAAGCCCAUGGCAUUGAUCAUAAUCGAAGCCACAGGAUAAUAAACUCUAGGAAUGCACUGCAA-GUCCAUUG (((....)))(((((((((..((((..((((.(((......)))))))....((..((((.(((((....))))).))))..))....................))))..)-)))))))) ( -34.30) >consensus _____CGGUUUAAUGGACUCCCAGUUAACGAUCCAAGUUGCUGGUCGUAAAGCCCAUGGCAUUGAUCAUAAUCGAUGCCACAGGAUAAUAAACUCUAGGAAUGCACUGCAA_GUCCAUUG ..........((((((((...((((...((.(((.(((((((........)))((.((((((((((....))))))))))..))......))))...))).)).))))....)))))))) (-29.88 = -30.32 + 0.44)


| Location | 4,422,091 – 4,422,205 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.85 |
| Mean single sequence MFE | -31.78 |
| Consensus MFE | -28.98 |
| Energy contribution | -29.22 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.982960 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4422091 114 - 22224390 CAGGUUUUAUGUCUGACACGUUCUUUUAGCUACCUUCCU------CGGUUUAAUGGACUCCCAGUUAACGAUCCAAGUUGCUGGUCGUAAAGCCCAUGGCAUUGAUCAUAAUCGAUGCCA ..(((((((((........((((..((((..(((.....------.))))))).))))..(((((.(((.......)))))))).)))))))))..((((((((((....)))))))))) ( -34.50) >DroSec_CAF1 24568 114 - 1 CAGGUUUUAUGUCUGACACGUUCUUUUGGCUACCUUCCU------CGGUUUAAUGGACUCCCAGUUAACGAUCCAAGUUGCUGGUCAUAAAGCCCAUGGCAUUGAUCAUAAUCGAUGCCA ..(((((((((........((((..((((..(((.....------.))))))).))))..(((((.(((.......)))))))).)))))))))..((((((((((....)))))))))) ( -34.50) >DroSim_CAF1 25880 114 - 1 CAGGUUUUAUGUCUGACACGUUCUUUUGGCUACCUUCCU------CGGUUUAAUGGACUCCCAGUUAACGAUCCAAGUUGCUGGUCGUAAAGCCCAUGGCAUUGAUCAUAAUCGAUGCCA ..(((((((((........((((..((((..(((.....------.))))))).))))..(((((.(((.......)))))))).)))))))))..((((((((((....)))))))))) ( -34.10) >DroEre_CAF1 25554 114 - 1 CAGGUUUUAUGUCUGACACGUUCUUUUAGCUACCUUCCU------CGGUUUAAUGGACCCGCAGUUAACGAUCCAAGUUGCUGGUCGUAAAGCCCAUGGCAUUGAUCAUAAUCGAAGGCG ..(((((((((.(......((((..((((..(((.....------.))))))).))))...((((.(((.......)))))))).)))))))))....((.(((((....)))))..)). ( -23.50) >DroYak_CAF1 25135 120 - 1 CAGGUUUUAUGUCUGACACGUUCUUUUAGCUACGUUCCUUAGCUACGGUUUAAUGGACUCCCAGUUAACGAUCCAAGUUGCUGGUCGUAAAGCCCAUGGCAUUGAUCAUAAUCGAAGCCA ..(((((((((((((((.(((......(((((.......)))))))))))....))))..(((((.(((.......))))))))..))))))))..((((.(((((....))))).)))) ( -32.30) >consensus CAGGUUUUAUGUCUGACACGUUCUUUUAGCUACCUUCCU______CGGUUUAAUGGACUCCCAGUUAACGAUCCAAGUUGCUGGUCGUAAAGCCCAUGGCAUUGAUCAUAAUCGAUGCCA ..(((((((((........((((....((((...............))))....))))..(((((.(((.......)))))))).)))))))))..((((((((((....)))))))))) (-28.98 = -29.22 + 0.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:10:54 2006