
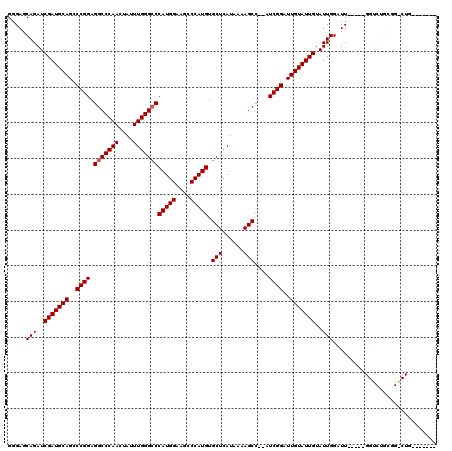
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,421,512 – 4,421,663 |
| Length | 151 |
| Max. P | 0.978835 |

| Location | 4,421,512 – 4,421,623 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.06 |
| Mean single sequence MFE | -31.88 |
| Consensus MFE | -26.16 |
| Energy contribution | -26.36 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.898225 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4421512 111 + 22224390 -------CAGCCCGCAGACCAAUCCAAUCCAAUACAAUACAAACCGAU--GGCUUUUAUGAGCACAUGGGCUUCCAUGGUCCCAAAUAGUUGGGCCUCCGGGCUGCAUCGAUCUGCUCCC -------(((((((.............................((.((--(((((....)))).))).)).......((.(((((....)))))))..)))))))............... ( -31.40) >DroSec_CAF1 24011 94 + 1 -------C-----------------AAUCCAAUACAAUACAAUCCGAU--GGCUUUUAUGAGCACAUGGGCUUCCAUGGGCCCAAAUAGUUGGGCCUCCGGGCUGCAUCGAUCUGCUCCC -------.-----------------..................((.((--(((((....)))).))).)).......((((((((....))))))))..(((..(((......))).))) ( -28.20) >DroSim_CAF1 25303 94 + 1 -------C-----------------AAUCCAAUACAAUACAAUCCGAU--GGCUUUUAUGAGCACAUGGGCUUCCAUGGGCCCAAAUAGUUGGGCCUCCGGGCUGCAUCGAUCUGCUCCC -------.-----------------..................((.((--(((((....)))).))).)).......((((((((....))))))))..(((..(((......))).))) ( -28.20) >DroEre_CAF1 24950 115 + 1 CAUCGCACAGUCCGCAGACC-----AAUCCAAUACAAUACAAUCCGAUCCGGCUUUUAUGAGCACAUGGGCUUCCAUGGGCCCAAAUAGUUGGGCCUCCGGGCUGCAUCGAUCUGCUCCC .((((..(((((((......-----..........................((((....)))).(((((....)))))(((((((....)))))))..)))))))...))))........ ( -35.00) >DroYak_CAF1 24542 109 + 1 ----GCACAGCCCGCAGACC-----AAUCCAAUACAAUACAAUCCGAU--GGCUUUUAUGAGCACAUGGGCUUCCAUGGGCCCAAAUAGUUGGGCCUCCGGGCUGCAUCGAUCUGCUCCC ----((((((((((......-----..................((.((--(((((....)))).))).)).......((((((((....)))))))).)))))))........))).... ( -36.60) >consensus _______CAG_CCGCAGACC_____AAUCCAAUACAAUACAAUCCGAU__GGCUUUUAUGAGCACAUGGGCUUCCAUGGGCCCAAAUAGUUGGGCCUCCGGGCUGCAUCGAUCUGCUCCC ...................................................((((....)))).(((((....)))))(((((((....)))))))...(((..(((......))).))) (-26.16 = -26.36 + 0.20)


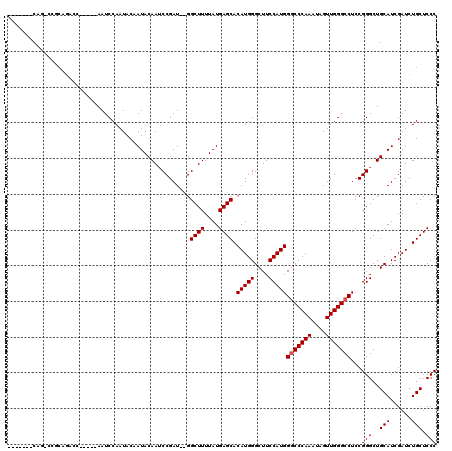
| Location | 4,421,512 – 4,421,623 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.06 |
| Mean single sequence MFE | -38.08 |
| Consensus MFE | -30.74 |
| Energy contribution | -30.94 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.785663 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4421512 111 - 22224390 GGGAGCAGAUCGAUGCAGCCCGGAGGCCCAACUAUUUGGGACCAUGGAAGCCCAUGUGCUCAUAAAAGCC--AUCGGUUUGUAUUGUAUUGGAUUGGAUUGGUCUGCGGGCUG------- .((.((((((((((.((..((((.(((((((....))))).))(((.(((((.(((.(((......))))--)).))))).)))....))))..)).))))))))))...)).------- ( -43.40) >DroSec_CAF1 24011 94 - 1 GGGAGCAGAUCGAUGCAGCCCGGAGGCCCAACUAUUUGGGCCCAUGGAAGCCCAUGUGCUCAUAAAAGCC--AUCGGAUUGUAUUGUAUUGGAUU-----------------G------- .....(((..(((((((..((((.(((((((....)))))))(((((....))))).(((......))).--.))))..)))))))..)))....-----------------.------- ( -32.30) >DroSim_CAF1 25303 94 - 1 GGGAGCAGAUCGAUGCAGCCCGGAGGCCCAACUAUUUGGGCCCAUGGAAGCCCAUGUGCUCAUAAAAGCC--AUCGGAUUGUAUUGUAUUGGAUU-----------------G------- .....(((..(((((((..((((.(((((((....)))))))(((((....))))).(((......))).--.))))..)))))))..)))....-----------------.------- ( -32.30) >DroEre_CAF1 24950 115 - 1 GGGAGCAGAUCGAUGCAGCCCGGAGGCCCAACUAUUUGGGCCCAUGGAAGCCCAUGUGCUCAUAAAAGCCGGAUCGGAUUGUAUUGUAUUGGAUU-----GGUCUGCGGACUGUGCGAUG ........((((.(((((.(((.(((((........(((((.(((((....))))).))))).........((((.(((........))).))))-----))))).))).))))))))). ( -40.90) >DroYak_CAF1 24542 109 - 1 GGGAGCAGAUCGAUGCAGCCCGGAGGCCCAACUAUUUGGGCCCAUGGAAGCCCAUGUGCUCAUAAAAGCC--AUCGGAUUGUAUUGUAUUGGAUU-----GGUCUGCGGGCUGUGC---- ..............((((((((.(((((........(((((.(((((....))))).)))))........--(((.(((........))).))).-----))))).))))))))..---- ( -41.50) >consensus GGGAGCAGAUCGAUGCAGCCCGGAGGCCCAACUAUUUGGGCCCAUGGAAGCCCAUGUGCUCAUAAAAGCC__AUCGGAUUGUAUUGUAUUGGAUU_____GGUCUGCGG_CUG_______ .....(((..(((((((..((((.(((((((....)))))))(((((....))))).(((......)))....))))..)))))))..)))............................. (-30.74 = -30.94 + 0.20)

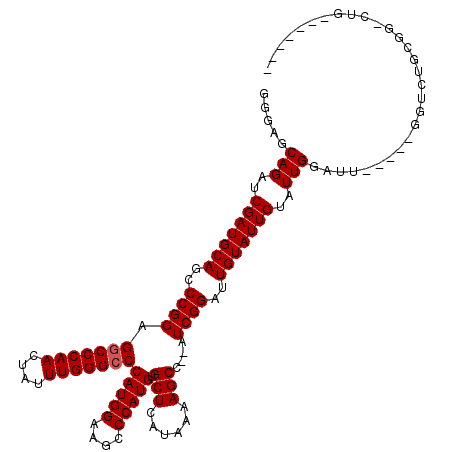
| Location | 4,421,545 – 4,421,663 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.19 |
| Mean single sequence MFE | -38.65 |
| Consensus MFE | -33.72 |
| Energy contribution | -33.92 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.586689 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4421545 118 + 22224390 AAACCGAU--GGCUUUUAUGAGCACAUGGGCUUCCAUGGUCCCAAAUAGUUGGGCCUCCGGGCUGCAUCGAUCUGCUCCCAGAAUUUUAAGUACUUGCCGAUCUGGCCGUCUGCCCAAAG ......((--(((((....)))).)))((((......((.(((((....)))))))...((((.((...((((.((....................)).))))..)).)))))))).... ( -34.05) >DroSec_CAF1 24027 107 + 1 AAUCCGAU--GGCUUUUAUGAGCACAUGGGCUUCCAUGGGCCCAAAUAGUUGGGCCUCCGGGCUGCAUCGAUCUGCUCCCAGAAUUUUAAGUACUUGCCGAUCUGGCCG----------- ........--((((....((.((((((((....)))))(((((((....)))))))((.(((..(((......))).))).))............)))))....)))).----------- ( -35.20) >DroSim_CAF1 25319 118 + 1 AAUCCGAU--GGCUUUUAUGAGCACAUGGGCUUCCAUGGGCCCAAAUAGUUGGGCCUCCGGGCUGCAUCGAUCUGCUCCCAGAAUUUUAAGUACUUGCCGAUCUGGCCGUCUGCCCAACG .....(((--((((....((.((((((((....)))))(((((((....)))))))((.(((..(((......))).))).))............)))))....)))))))......... ( -40.20) >DroEre_CAF1 24985 120 + 1 AAUCCGAUCCGGCUUUUAUGAGCACAUGGGCUUCCAUGGGCCCAAAUAGUUGGGCCUCCGGGCUGCAUCGAUCUGCUCCCAGAAUUUUAAGUACUUGCCGAUCUGGCCGGCUGCCCAAAG ...........((((....))))...(((((..((..((((((((....))))))))...(((((.((((.((((....)))).......((....)))))).)))))))..)))))... ( -41.60) >DroYak_CAF1 24573 117 + 1 AAUCCGAU--GGCUUUUAUGAGCACAUGGGCUUCCAUGGGCCCAAAUAGUUGGGCCUCCGGGCUGCAUCGAUCUGCUCCCAGAAUUUUAAGUACUUGCCGAUCUGGCCGGCUGCCCAAA- ......((--(((((....)))).)))((((..((..((((((((....))))))))...(((((.((((.((((....)))).......((....)))))).)))))))..))))...- ( -42.20) >consensus AAUCCGAU__GGCUUUUAUGAGCACAUGGGCUUCCAUGGGCCCAAAUAGUUGGGCCUCCGGGCUGCAUCGAUCUGCUCCCAGAAUUUUAAGUACUUGCCGAUCUGGCCGGCUGCCCAAAG ..........((((....((.((((((((....)))))(((((((....)))))))((.(((..(((......))).))).))............)))))....))))............ (-33.72 = -33.92 + 0.20)


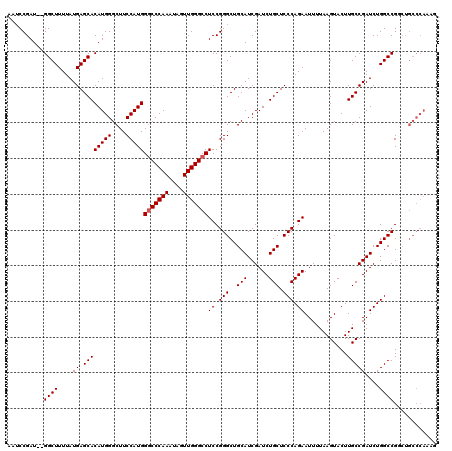
| Location | 4,421,545 – 4,421,663 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.19 |
| Mean single sequence MFE | -42.12 |
| Consensus MFE | -35.42 |
| Energy contribution | -36.82 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.978835 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4421545 118 - 22224390 CUUUGGGCAGACGGCCAGAUCGGCAAGUACUUAAAAUUCUGGGAGCAGAUCGAUGCAGCCCGGAGGCCCAACUAUUUGGGACCAUGGAAGCCCAUGUGCUCAUAAAAGCC--AUCGGUUU ..((((((.....(((.....)))............(((((((.(((......)))..))))))))))))).....((((..(((((....)))))..))))...(((((--...))))) ( -39.40) >DroSec_CAF1 24027 107 - 1 -----------CGGCCAGAUCGGCAAGUACUUAAAAUUCUGGGAGCAGAUCGAUGCAGCCCGGAGGCCCAACUAUUUGGGCCCAUGGAAGCCCAUGUGCUCAUAAAAGCC--AUCGGAUU -----------.(((...((.((((............((((((.(((......)))..))))))(((((((....)))))))(((((....))))))))).))....)))--........ ( -38.30) >DroSim_CAF1 25319 118 - 1 CGUUGGGCAGACGGCCAGAUCGGCAAGUACUUAAAAUUCUGGGAGCAGAUCGAUGCAGCCCGGAGGCCCAACUAUUUGGGCCCAUGGAAGCCCAUGUGCUCAUAAAAGCC--AUCGGAUU .(((((((.....(((.....)))............(((((((.(((......)))..))))))))))))))....(((((.(((((....))))).)))))........--........ ( -44.30) >DroEre_CAF1 24985 120 - 1 CUUUGGGCAGCCGGCCAGAUCGGCAAGUACUUAAAAUUCUGGGAGCAGAUCGAUGCAGCCCGGAGGCCCAACUAUUUGGGCCCAUGGAAGCCCAUGUGCUCAUAAAAGCCGGAUCGGAUU .(((((....(((((...((.((((............((((((.(((......)))..))))))(((((((....)))))))(((((....))))))))).))....))))).))))).. ( -45.10) >DroYak_CAF1 24573 117 - 1 -UUUGGGCAGCCGGCCAGAUCGGCAAGUACUUAAAAUUCUGGGAGCAGAUCGAUGCAGCCCGGAGGCCCAACUAUUUGGGCCCAUGGAAGCCCAUGUGCUCAUAAAAGCC--AUCGGAUU -.((((((.(((((.....)))))............(((((((.(((......)))..))))))))))))).....(((((.(((((....))))).)))))........--........ ( -43.50) >consensus CUUUGGGCAGACGGCCAGAUCGGCAAGUACUUAAAAUUCUGGGAGCAGAUCGAUGCAGCCCGGAGGCCCAACUAUUUGGGCCCAUGGAAGCCCAUGUGCUCAUAAAAGCC__AUCGGAUU ..((((((.....(((.....)))............(((((((.(((......)))..))))))))))))).....(((((.(((((....))))).))))).................. (-35.42 = -36.82 + 1.40)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:10:50 2006