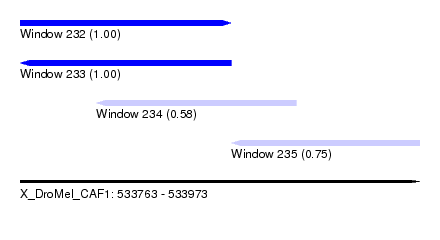
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 533,763 – 533,973 |
| Length | 210 |
| Max. P | 0.999578 |

| Location | 533,763 – 533,874 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.89 |
| Mean single sequence MFE | -30.60 |
| Consensus MFE | -28.76 |
| Energy contribution | -28.84 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.70 |
| SVM RNA-class probability | 0.996471 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 533763 111 + 22224390 AUUUUGGCUCAAUUUUUUCCCACUCAGGUUUGAACUAUCAAAGGCCAAAGGGGCAUUGGGUUUCAACAGAGAUGUGUUUUGCCCCAUUUGGUCCUUUCUGCAUUUUCAAGC--------- ...........................(((((((.....((((((((((((((((.((.(((((....))))).))...)))))).))))).))))).......)))))))--------- ( -31.00) >DroSec_CAF1 21745 111 + 1 AUUUUGGCUCAAUUUUUUCCCACCCAGGUUUGAACUAUCAAAGGCCAAAGGGGCAUUGGGUUACAAAAGAGAUGUGUUUUGCCCCAUUUGGUCCUUUCUGCAUUUUCGAGC--------- ......((((......(((..((....))..))).....((((((((((((((((......((((.......))))...)))))).))))).)))))..........))))--------- ( -29.60) >DroSim_CAF1 21906 111 + 1 AUUUUGGCUCAAUUUUUUCCCACUCAGGUUUGAACUAUCAAAGGCCAAAGGGGCAUUGGGUUACAAAAGAGAUGUGUUUUGCCCCAUUUGGUCCUUUCUGCAUUUUCAAGC--------- ...........................(((((((.....((((((((((((((((......((((.......))))...)))))).))))).))))).......)))))))--------- ( -27.80) >DroEre_CAF1 25041 111 + 1 AUUUUGGCUCAAUUUUUUCCCACUCAGGUUUGAACUAUCAAAGGCCAAAGGGGCAUUGGGUUUCAAAAGAGAUGUGUUUUGCCCCAUUUGGUCCUUUCUGCAUUUUCAAGC--------- ...........................(((((((.....((((((((((((((((.((.(((((....))))).))...)))))).))))).))))).......)))))))--------- ( -31.00) >DroYak_CAF1 19366 120 + 1 AUUUUGGCUCAAUUUUUUGCCACUCAGGUUUGAACUAUCAAAGGCCAAAGGGGCAUUGGGUUUCAAAAGAGAUGUGUGCUGCCCCAUUUGGUCCUUUCUGCAUUUUCAAGUUUUCCAGCU ....((((..........))))....((...(((((...((((((((((((((((.((.(((((....))))).))...)))))).))))).))))).((......))))))).)).... ( -33.60) >consensus AUUUUGGCUCAAUUUUUUCCCACUCAGGUUUGAACUAUCAAAGGCCAAAGGGGCAUUGGGUUUCAAAAGAGAUGUGUUUUGCCCCAUUUGGUCCUUUCUGCAUUUUCAAGC_________ ...........................(((((((.....((((((((((((((((.((.(((((....))))).))...)))))).))))).))))).......)))))))......... (-28.76 = -28.84 + 0.08)


| Location | 533,763 – 533,874 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.89 |
| Mean single sequence MFE | -31.00 |
| Consensus MFE | -27.32 |
| Energy contribution | -27.76 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.88 |
| SVM decision value | 3.74 |
| SVM RNA-class probability | 0.999578 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 533763 111 - 22224390 ---------GCUUGAAAAUGCAGAAAGGACCAAAUGGGGCAAAACACAUCUCUGUUGAAACCCAAUGCCCCUUUGGCCUUUGAUAGUUCAAACCUGAGUGGGAAAAAAUUGAGCCAAAAU ---------(((..(........(((((.(((((.((((((.......((......)).......))))))))))))))))...........(((....)))......)..)))...... ( -30.34) >DroSec_CAF1 21745 111 - 1 ---------GCUCGAAAAUGCAGAAAGGACCAAAUGGGGCAAAACACAUCUCUUUUGUAACCCAAUGCCCCUUUGGCCUUUGAUAGUUCAAACCUGGGUGGGAAAAAAUUGAGCCAAAAU ---------((((((........(((((.(((((.((((((....(((.......))).......)))))))))))))))).....(((..((....))..)))....))))))...... ( -33.00) >DroSim_CAF1 21906 111 - 1 ---------GCUUGAAAAUGCAGAAAGGACCAAAUGGGGCAAAACACAUCUCUUUUGUAACCCAAUGCCCCUUUGGCCUUUGAUAGUUCAAACCUGAGUGGGAAAAAAUUGAGCCAAAAU ---------(((..(........(((((.(((((.((((((....(((.......))).......))))))))))))))))...........(((....)))......)..)))...... ( -29.60) >DroEre_CAF1 25041 111 - 1 ---------GCUUGAAAAUGCAGAAAGGACCAAAUGGGGCAAAACACAUCUCUUUUGAAACCCAAUGCCCCUUUGGCCUUUGAUAGUUCAAACCUGAGUGGGAAAAAAUUGAGCCAAAAU ---------(((..(........(((((.(((((.((((((.......((......)).......))))))))))))))))...........(((....)))......)..)))...... ( -30.34) >DroYak_CAF1 19366 120 - 1 AGCUGGAAAACUUGAAAAUGCAGAAAGGACCAAAUGGGGCAGCACACAUCUCUUUUGAAACCCAAUGCCCCUUUGGCCUUUGAUAGUUCAAACCUGAGUGGCAAAAAAUUGAGCCAAAAU .(((.((....(((..(...((((((((.(((((.((((((.......((......)).......))))))))))))))))((....))....)))..)..)))....)).)))...... ( -31.74) >consensus _________GCUUGAAAAUGCAGAAAGGACCAAAUGGGGCAAAACACAUCUCUUUUGAAACCCAAUGCCCCUUUGGCCUUUGAUAGUUCAAACCUGAGUGGGAAAAAAUUGAGCCAAAAU .........((((((........(((((.(((((.((((((.......((......)).......))))))))))))))))...........((.....)).......))))))...... (-27.32 = -27.76 + 0.44)

| Location | 533,803 – 533,908 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.63 |
| Mean single sequence MFE | -29.08 |
| Consensus MFE | -23.49 |
| Energy contribution | -23.89 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.584848 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 533803 105 - 22224390 GUGUACGAACGGACAGCCGACAGCUCAAUUCUCA---------------GCUUGAAAAUGCAGAAAGGACCAAAUGGGGCAAAACACAUCUCUGUUGAAACCCAAUGCCCCUUUGGCCUU .((((....(((....)))..((((........)---------------)))......))))..((((.(((((.((((((.......((......)).......))))))))))))))) ( -28.14) >DroSec_CAF1 21785 105 - 1 GUGUUCGGACGGACAGCCGACAGCUCAAUUCUCA---------------GCUCGAAAAUGCAGAAAGGACCAAAUGGGGCAAAACACAUCUCUUUUGUAACCCAAUGCCCCUUUGGCCUU ((.((((..(((....)))..((((........)---------------)))))))...))...((((.(((((.((((((....(((.......))).......))))))))))))))) ( -29.30) >DroSim_CAF1 21946 105 - 1 GUGUACGGACGGACAGCCGACAGCUCAAUUCUCA---------------GCUUGAAAAUGCAGAAAGGACCAAAUGGGGCAAAACACAUCUCUUUUGUAACCCAAUGCCCCUUUGGCCUU .(((.(((.(.....)))))))((....(((...---------------....)))...))...((((.(((((.((((((....(((.......))).......))))))))))))))) ( -27.90) >DroEre_CAF1 25081 101 - 1 --GUGU--ACGGACAGCCGACAGCUCAAUUCUCA---------------GCUUGAAAAUGCAGAAAGGACCAAAUGGGGCAAAACACAUCUCUUUUGAAACCCAAUGCCCCUUUGGCCUU --.(((--.(((....))))))((....(((...---------------....)))...))...((((.(((((.((((((.......((......)).......))))))))))))))) ( -28.14) >DroYak_CAF1 19406 118 - 1 GUGUGU--GCGGACAGCCGACAGCUCAAUUCUCAAUUCUCAGCUGGAAAACUUGAAAAUGCAGAAAGGACCAAAUGGGGCAGCACACAUCUCUUUUGAAACCCAAUGCCCCUUUGGCCUU .(((((--.(((....))).(((((...............)))))............)))))..((((.(((((.((((((.......((......)).......))))))))))))))) ( -31.90) >consensus GUGUACG_ACGGACAGCCGACAGCUCAAUUCUCA_______________GCUUGAAAAUGCAGAAAGGACCAAAUGGGGCAAAACACAUCUCUUUUGAAACCCAAUGCCCCUUUGGCCUU .........(((....)))...((....(((......................)))...))...((((.(((((.((((((.......((......)).......))))))))))))))) (-23.49 = -23.89 + 0.40)

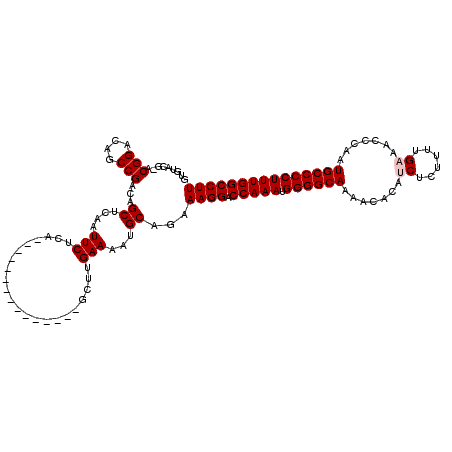
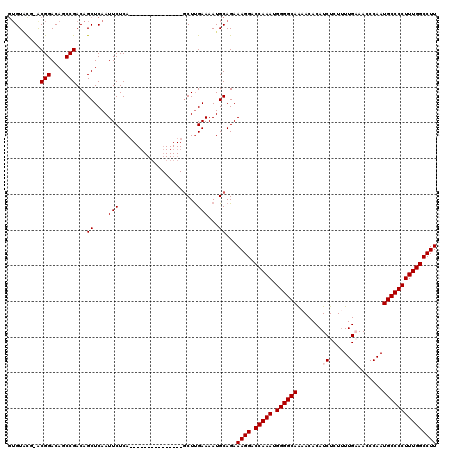
| Location | 533,874 – 533,973 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 86.87 |
| Mean single sequence MFE | -30.61 |
| Consensus MFE | -22.48 |
| Energy contribution | -23.22 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.753293 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 533874 99 - 22224390 ---------------AUGACCCAGAGAAUACCCUUUCGUACACACACUGGCCUAGUUGCCCACACAGAUGCACGCAUUGUGUGUACGAACGGACAGCCGACAGCUCAAUUCUCA ---------------........((((((..((.(((((((((((...(((......)))......((((....))))))))))))))).))..(((.....)))..)))))). ( -32.90) >DroSec_CAF1 21856 112 - 1 GUCCCAGAGAUUUUGAUGGCCCA--GAAUACCCUUUCGUACACACACUGGCCUAGUUGCCCACACAGAUGCACGCAUUGUGUGUUCGGACGGACAGCCGACAGCUCAAUUCUCA ......((((..((((.((((..--(((......)))((....))...))))..((((..(((((((.((....))))))))).((((.(.....))))))))))))).)))). ( -30.40) >DroSim_CAF1 22017 112 - 1 GUCCCACAGAUUUUGAUGGCCCA--GAAUACCCUUUCGUACACACACUGGCCUAGUUGCCCACACAGAUGCACGCAUUGUGUGUACGGACGGACAGCCGACAGCUCAAUUCUCA ((((.....((((((......))--)))).....(((((((((((...(((......)))......((((....))))))))))))))).))))(((.....)))......... ( -31.10) >DroEre_CAF1 25152 108 - 1 GUCUCAGAGAUUUUGAUGGCCCA--GAAUACCCUUUCACACACACACUGGCCUAGUUGCCCACACACAUGCACGCAUUGU--GUGU--ACGGACAGCCGACAGCUCAAUUCUCA ......((((..((((.((((.(--(....................))))))..((((...(((((((((....)).)))--))))--.(((....))).)))))))).)))). ( -28.05) >consensus GUCCCAGAGAUUUUGAUGGCCCA__GAAUACCCUUUCGUACACACACUGGCCUAGUUGCCCACACAGAUGCACGCAUUGUGUGUACGGACGGACAGCCGACAGCUCAAUUCUCA .........................((((..((.(((((((((((...(((......)))......((((....))))))))))))))).))..(((.....)))..))))... (-22.48 = -23.22 + 0.75)

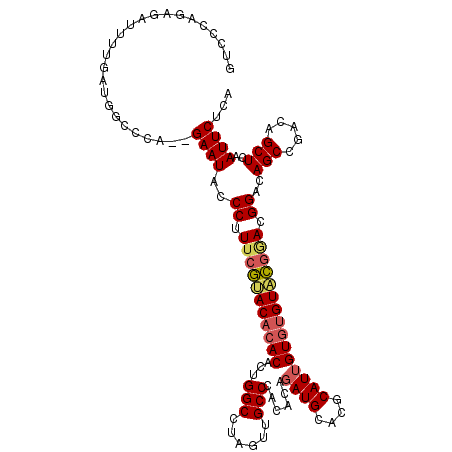
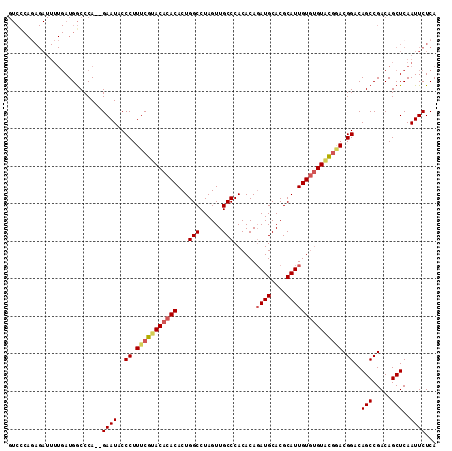
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:37:11 2006