
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,421,272 – 4,421,392 |
| Length | 120 |
| Max. P | 0.808100 |

| Location | 4,421,272 – 4,421,392 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.43 |
| Mean single sequence MFE | -28.32 |
| Consensus MFE | -25.18 |
| Energy contribution | -24.94 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.808100 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
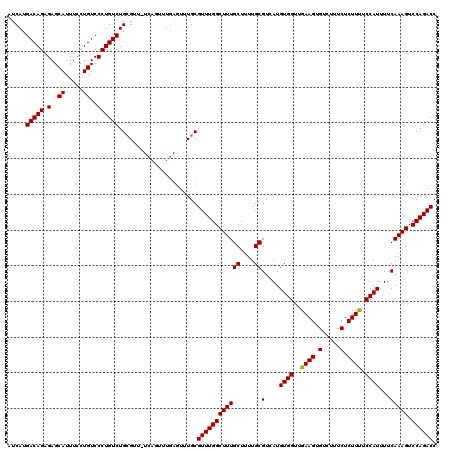
>X_DroMel_CAF1 4421272 120 + 22224390 AUCAUGACAGAGAGCAUUUCCUGUCCCUGUCUGCGUUUUCAGUUUGAGUUUGCGUUUGGCUUUGCUUUUGCGUCAUGUGGUUAAAGUGUCUUUCUCUUUUCCAUUUUCAAAGUCCAGACC .....(((((.(..((.....))..)))))).(((..(((.....)))..)))((((((((((((....))(....((((..((((.(.....).)))).))))...))))).)))))). ( -29.80) >DroSec_CAF1 23772 119 + 1 AUCAUGACAGAGAGCAUUUCCUGUCCCUGUCUGCGUU-UCAGUUUGAGUUUGCGUUUGGCUUUGCUUUUGCGUCAUGUGGUUAAAGUGUCUUUCUCUUUUCCAUUUUCAAAGUCCAGACC .....(((((.(..((.....))..)))))).(((.(-((.....)))..)))((((((((((((....))(....((((..((((.(.....).)))).))))...))))).)))))). ( -27.70) >DroSim_CAF1 25064 119 + 1 AUCAUGACAGAGAGCAUUUCCUGUCCCUGUCUGCGUU-UCAGUUUGAGUUUGCGUUUGGCUUUGCUUUUGCGUCAUGUGGUUGAAGUGUCUUUCUCUUUUCCAUUUUCAAAGUCCAGACC .....(((((.(..((.....))..)))))).(((.(-((.....)))..)))((((((((((((....))(....((((..((((.(.....).)))).))))...))))).)))))). ( -28.10) >DroEre_CAF1 24721 113 + 1 AUCAUGACAGAGAGCAUUUCCUGUCCCUGUCUGGG-------UUUGAGUUUGCGUUUGGCUUUGCUUUUGCGUCAUGUGGUUGAAGUGUCUUUCUCUUUUCCAUUUUCAAAGUCCAGACC .....(((((.(((...))))))))...(((((((-------((((((...((((..(((...)))...))))...((((..((((.(.....).)))).)))).)))))).))))))). ( -30.30) >DroYak_CAF1 24329 107 + 1 AUCAUGACAGAGAGCAUUUCCUGUCCCUGUCUGCG-------------UUUGCGUUUGGCUUUGCUUUUGCGUUAUGUGGUUGAAGUGUCUUUCUCUUUUCCAUUUUCAAAGUCCAGACC .....(((((.(..((.....))..)))))).((.-------------...))((((((((((((....))(....((((..((((.(.....).)))).))))...))))).)))))). ( -25.70) >consensus AUCAUGACAGAGAGCAUUUCCUGUCCCUGUCUGCGUU_UCAGUUUGAGUUUGCGUUUGGCUUUGCUUUUGCGUCAUGUGGUUGAAGUGUCUUUCUCUUUUCCAUUUUCAAAGUCCAGACC .....(((((.(..((.....))..))))))......................((((((((((((....))(....((((..((((.(.....).)))).))))...))))).)))))). (-25.18 = -24.94 + -0.24)


| Location | 4,421,272 – 4,421,392 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.43 |
| Mean single sequence MFE | -23.32 |
| Consensus MFE | -21.90 |
| Energy contribution | -21.90 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.07 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.559507 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
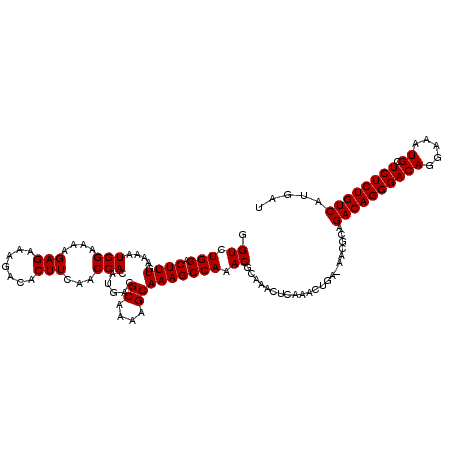
>X_DroMel_CAF1 4421272 120 - 22224390 GGUCUGGACUUUGAAAAUGGAAAAGAGAAAGACACUUUAACCACAUGACGCAAAAGCAAAGCCAAACGCAAACUCAAACUGAAAACGCAGACAGGGACAGGAAAUGCUCUCUGUCAUGAU .((.(((.((((.....(((...((((.......))))..)))......((....))))))))).))((....((.....))....)).((((((((((.....)).))))))))..... ( -23.90) >DroSec_CAF1 23772 119 - 1 GGUCUGGACUUUGAAAAUGGAAAAGAGAAAGACACUUUAACCACAUGACGCAAAAGCAAAGCCAAACGCAAACUCAAACUGA-AACGCAGACAGGGACAGGAAAUGCUCUCUGUCAUGAU .((.(((.((((.....(((...((((.......))))..)))......((....))))))))).))((....((.....))-...)).((((((((((.....)).))))))))..... ( -23.50) >DroSim_CAF1 25064 119 - 1 GGUCUGGACUUUGAAAAUGGAAAAGAGAAAGACACUUCAACCACAUGACGCAAAAGCAAAGCCAAACGCAAACUCAAACUGA-AACGCAGACAGGGACAGGAAAUGCUCUCUGUCAUGAU .((.(((.((((.....(((....(((.......)))...)))......((....))))))))).))((....((.....))-...)).((((((((((.....)).))))))))..... ( -22.50) >DroEre_CAF1 24721 113 - 1 GGUCUGGACUUUGAAAAUGGAAAAGAGAAAGACACUUCAACCACAUGACGCAAAAGCAAAGCCAAACGCAAACUCAAA-------CCCAGACAGGGACAGGAAAUGCUCUCUGUCAUGAU (((...((.((((....(((......(((......)))...........((....))....)))....)))).))..)-------))..((((((((((.....)).))))))))..... ( -23.50) >DroYak_CAF1 24329 107 - 1 GGUCUGGACUUUGAAAAUGGAAAAGAGAAAGACACUUCAACCACAUAACGCAAAAGCAAAGCCAAACGCAAA-------------CGCAGACAGGGACAGGAAAUGCUCUCUGUCAUGAU .((.(((.((((.....(((....(((.......)))...)))......((....))))))))).))((...-------------.)).((((((((((.....)).))))))))..... ( -23.20) >consensus GGUCUGGACUUUGAAAAUGGAAAAGAGAAAGACACUUCAACCACAUGACGCAAAAGCAAAGCCAAACGCAAACUCAAACUGA_AACGCAGACAGGGACAGGAAAUGCUCUCUGUCAUGAU .((.(((.((((.....(((....(((.......)))...)))......((....))))))))).))......................((((((((((.....)).))))))))..... (-21.90 = -21.90 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:10:46 2006