
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,417,350 – 4,417,620 |
| Length | 270 |
| Max. P | 0.999020 |

| Location | 4,417,350 – 4,417,463 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.43 |
| Mean single sequence MFE | -21.26 |
| Consensus MFE | -19.04 |
| Energy contribution | -19.36 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.980181 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4417350 113 + 22224390 AUGGGCAAAGUUGCCAAAUGCAACGAGCUAACAACCGCAUCUAGUGGUGCAUCAUGCAAUCUUCCUCUUCAAUUUCCAUUUA-------CCAUUUGCCGCAUUCAUAAAACAAACAAAAU ((((((((((((((.....)))))(((......(((((.....)))))((.....)).......)))...............-------...)))))).))).................. ( -23.80) >DroSec_CAF1 19771 120 + 1 AUGGACAAAGUUGCCAAAUGCAACGAGCUAACAACCGCAUCUAGUGGUGCAUCAUGCAAUCUUCCUCUUCAACUUCCAUUUUCCAUUUUCCUUUUGCCGCAUUCAUAAAAUAAACAAAAU ..((.(((((((((.....)))))(((......(((((.....)))))((.....)).......))).........................))))))...................... ( -19.30) >DroSim_CAF1 21217 120 + 1 AUGGCCAAAGUUGCCAAAUGCAACGAGCUAACAACCGCAUCUAGUGGUGCAUCAUGCAAUCUUCCUCUUCAACUUCCAUUUUCCAUUUUCCAUUUGCUGCAUUCAUAAAACAAACAAAAU .((((.......))))((((((.((((......(((((.....)))))((.....))...................................)))).))))))................. ( -18.80) >DroEre_CAF1 20820 113 + 1 AUGGGCAAAGUUGCCAAAUGCAACGAGCAAACAACCGCAUCUAGUGGUGCAUCAUGCAAUCUUCCUCUUCAAUUUCCAUUUU-------CCAUUUGCCGCAGUCAUAAAACAAACAAAAU .(((((((((((((.....)))))(((......(((((.....)))))((.....)).......)))...............-------...)))))).))................... ( -23.70) >DroYak_CAF1 20482 113 + 1 AUGGGCAGAGUUGCCAAAUGCAGCGACCAAACAACCGCAUCUAGUGGUGCAUCAUGCAAUCUUCCUCUUCAAUUUCCAUUUU-------CCAAUUGCCGCAUUCAAAAAACAAACAAAAU ...(((......))).(((((.((((.......(((((.....)))))((.....)).........................-------....)))).)))))................. ( -20.70) >consensus AUGGGCAAAGUUGCCAAAUGCAACGAGCUAACAACCGCAUCUAGUGGUGCAUCAUGCAAUCUUCCUCUUCAAUUUCCAUUUU_______CCAUUUGCCGCAUUCAUAAAACAAACAAAAU .(((((((((((((.....)))))(((......(((((.....)))))((.....)).......))).........................)))))).))................... (-19.04 = -19.36 + 0.32)


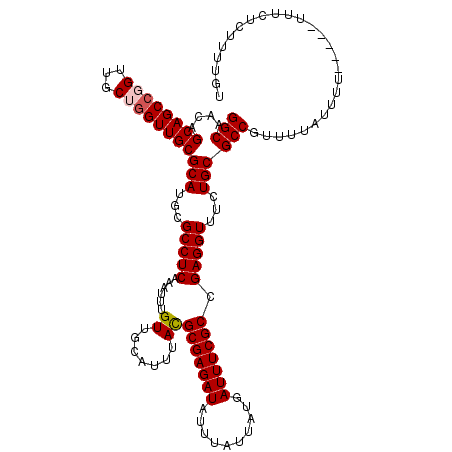
| Location | 4,417,430 – 4,417,543 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.51 |
| Mean single sequence MFE | -22.27 |
| Consensus MFE | -22.34 |
| Energy contribution | -22.40 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.24 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.693501 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4417430 113 - 22224390 UUUGUUGCAUUUACGCGAGAUAUUUAUUAUGAUUUCGCCGAGGUUUCUGCGCCGUUUUAUUUUUCAUUUUUCUCUUUUGUAUUUUGUUUGUUUUAUGAAUGCGGCAAAUGG-------UA (((((((((((((.(((((((..........)))))))...(((......)))..........................................)))))))))))))...-------.. ( -22.70) >DroSec_CAF1 19851 113 - 1 UUUGUUGCAUUUAUGCGAGAUAUUUAUUAUGAUUUCGCCGAGGUUUCUGCGCCGUUUUAUU-------UCUCUCUUUUGUAUUUUGUUUAUUUUAUGAAUGCGGCAAAAGGAAAAUGGAA (((((((((((((((((((((..........)))))))...(((......)))........-------..........................))))))))))))))............ ( -23.80) >DroSim_CAF1 21297 113 - 1 UUUGUUGCAUUUACGCGAGAUAUUUAUUAUGAUUUCGCCGAGGUUUCUGCGCCGUUUUAUU-------UCUCUCUUUUGUAUUUUGUUUGUUUUAUGAAUGCAGCAAAUGGAAAAUGGAA (((((((((((((.(((((((..........)))))))...(((......)))........-------...........................)))))))))))))............ ( -23.50) >DroEre_CAF1 20900 108 - 1 UUUGUUGCAUUUACGCGAGAUAUUUAUUAUGAUUUCGCCGAGGUUUCUGCGCCGUUUUAUUUU-----UUUCUCUUUUGUAUUUUGUUUGUUUUAUGACUGCGGCAAAUGG-------AA (((((((((..((((((((((..........)))))))...(((......)))..........-----..........)))........(((....))))))))))))...-------.. ( -19.10) >consensus UUUGUUGCAUUUACGCGAGAUAUUUAUUAUGAUUUCGCCGAGGUUUCUGCGCCGUUUUAUU_______UCUCUCUUUUGUAUUUUGUUUGUUUUAUGAAUGCGGCAAAUGG_______AA (((((((((((((.(((((((..........)))))))...(((......)))..........................................)))))))))))))............ (-22.34 = -22.40 + 0.06)



| Location | 4,417,463 – 4,417,583 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.69 |
| Mean single sequence MFE | -33.28 |
| Consensus MFE | -31.28 |
| Energy contribution | -31.72 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.94 |
| SVM decision value | 3.33 |
| SVM RNA-class probability | 0.999020 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4417463 120 - 22224390 GGCAGCAGCAGCCGGUUGCUAGUUGCGCAUGCGCCUCAAAUUUGUUGCAUUUACGCGAGAUAUUUAUUAUGAUUUCGCCGAGGUUUCUGCGCCGUUUUAUUUUUCAUUUUUCUCUUUUGU (((.((((((((.....))).)))))(((.(.(((((......((.......))(((((((..........))))))).))))).).))))))........................... ( -32.30) >DroSec_CAF1 19891 113 - 1 GGCAACAGCAGCCGGUUGCAGGUUGCGCAUGCGCCUCAAAUUUGUUGCAUUUAUGCGAGAUAUUUAUUAUGAUUUCGCCGAGGUUUCUGCGCCGUUUUAUU-------UCUCUCUUUUGU (((....((((((.......))))))(((.(.(((((.......(((((....)))))((.(((......))).))...))))).).))))))........-------............ ( -31.70) >DroSim_CAF1 21337 113 - 1 GGCAACAGCAGCCGGUUGCAGGUUGCGCAUGCGCCUCAAAUUUGUUGCAUUUACGCGAGAUAUUUAUUAUGAUUUCGCCGAGGUUUCUGCGCCGUUUUAUU-------UCUCUCUUUUGU (((....((((((.......))))))(((.(.(((((......((.......))(((((((..........))))))).))))).).))))))........-------............ ( -32.90) >DroEre_CAF1 20933 115 - 1 GGCAGCAGCAGCCGGUUGCUGGUUGCGCAUGCGCCUCAAAUUUGUUGCAUUUACGCGAGAUAUUUAUUAUGAUUUCGCCGAGGUUUCUGCGCCGUUUUAUUUU-----UUUCUCUUUUGU (((....((((((((...))))))))(((.(.(((((......((.......))(((((((..........))))))).))))).).))))))..........-----............ ( -34.40) >DroYak_CAF1 20595 115 - 1 GGCAACAGCAGCCGGUUGCUGGUUGCGCAUGCGCCUCAAAUUUGUUGCAUUUACGCGAGAUAUUUAUUAUGAUUUCGCCGAGGUUUUUGCGCCGUUUUAUUUU-----UUUCUCUUUUGU (((....((((((((...))))))))(((...(((((......((.......))(((((((..........))))))).)))))...))))))..........-----............ ( -35.10) >consensus GGCAACAGCAGCCGGUUGCUGGUUGCGCAUGCGCCUCAAAUUUGUUGCAUUUACGCGAGAUAUUUAUUAUGAUUUCGCCGAGGUUUCUGCGCCGUUUUAUUUU_____UUUCUCUUUUGU (((....((((((((...))))))))(((...(((((......((.......))(((((((..........))))))).)))))...))))))........................... (-31.28 = -31.72 + 0.44)


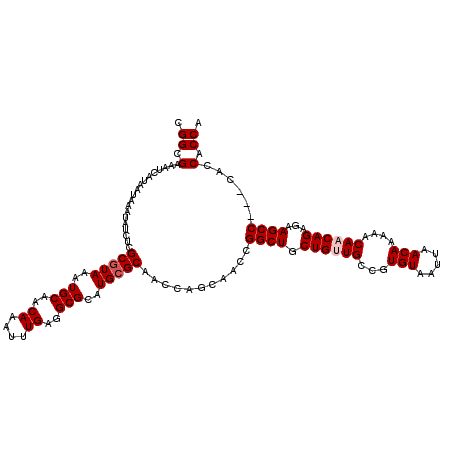
| Location | 4,417,503 – 4,417,620 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.47 |
| Mean single sequence MFE | -27.58 |
| Consensus MFE | -24.92 |
| Energy contribution | -25.52 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.04 |
| SVM RNA-class probability | 0.986327 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4417503 117 + 22224390 CGGCGAAAUCAUAAUAAAUAUCUCGCGUAAAUGCAACAAAUUUGAGGCGCAUGCGCAACUAGCAACCGGCUGCUGCUGCCGUGUAAUUAACAAAAACAACAGAGAAGCC---CACCACCA .((((....)..........(((((((((..(((..((....))..)))..)))))........((((((.......)))).)).................)))).)))---........ ( -26.20) >DroSec_CAF1 19924 120 + 1 CGGCGAAAUCAUAAUAAAUAUCUCGCAUAAAUGCAACAAAUUUGAGGCGCAUGCGCAACCUGCAACCGGCUGCUGUUGCCGUGUAAUUAACAAAAACAACAGAGAAGCCAACCACCACCA .((((....)..........(((((((....))).......((((...(((((.(((((..(((......))).))))))))))..))))...........)))).)))........... ( -27.10) >DroSim_CAF1 21370 120 + 1 CGGCGAAAUCAUAAUAAAUAUCUCGCGUAAAUGCAACAAAUUUGAGGCGCAUGCGCAACCUGCAACCGGCUGCUGUUGCCGUGUAAUUAACAAAAACAACAGAGAAGCCAACCACCACCA .((.(...................(((((..(((..((....))..)))..)))))...........((((.((((((...(((.....)))....))))))...))))...).)).... ( -28.80) >DroEre_CAF1 20968 117 + 1 CGGCGAAAUCAUAAUAAAUAUCUCGCGUAAAUGCAACAAAUUUGAGGCGCAUGCGCAACCAGCAACCGGCUGCUGCUGCCGUGUAAUUAACAAAAACAACAGAGAAGCC---CACCACCA .((((....)..........((((((.(((((.......)))))..))(((((.(((..(((((......))))).)))))))).................)))).)))---........ ( -26.30) >DroYak_CAF1 20630 117 + 1 CGGCGAAAUCAUAAUAAAUAUCUCGCGUAAAUGCAACAAAUUUGAGGCGCAUGCGCAACCAGCAACCGGCUGCUGUUGCCGUGUAAUUAACAAAAACAACAGAGAAGCC---CAUCACCA .((.((..................(((((..(((..((....))..)))..)))))...........((((.((((((...(((.....)))....))))))...))))---..)).)). ( -29.50) >consensus CGGCGAAAUCAUAAUAAAUAUCUCGCGUAAAUGCAACAAAUUUGAGGCGCAUGCGCAACCAGCAACCGGCUGCUGUUGCCGUGUAAUUAACAAAAACAACAGAGAAGCC___CACCACCA .((.(...................(((((..(((..((....))..)))..)))))...........((((.((((((...(((.....)))....))))))...))))......).)). (-24.92 = -25.52 + 0.60)


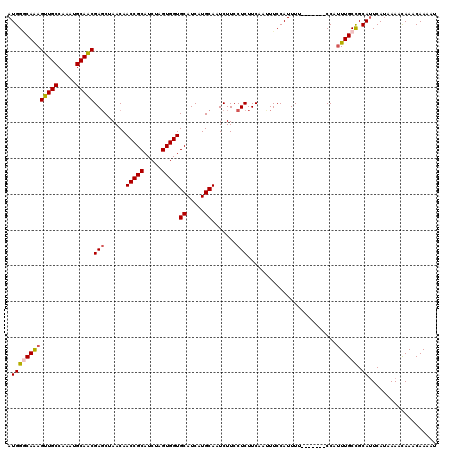
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:10:38 2006