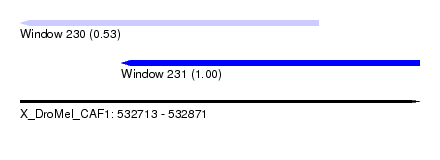
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 532,713 – 532,871 |
| Length | 158 |
| Max. P | 0.999889 |

| Location | 532,713 – 532,831 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.45 |
| Mean single sequence MFE | -22.36 |
| Consensus MFE | -18.64 |
| Energy contribution | -19.24 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.83 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.530709 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 532713 118 - 22224390 CAAAUUAAUUUCAAUUUGCCGUAAAUUAUUUGGCGCAUUC--UCCCAGCCAUGUAGAUUGCACAGCAAUUAACUGCAAAACAUUAACCGCAAACUAAGCGUAAGGUAAACAAGUAAUCGA ..............((((((((((.((((.((((......--.....)))).)))).))))...(((......)))...........(((.......)))...))))))........... ( -21.30) >DroSec_CAF1 20718 117 - 1 CAAAUUAAUUUCAAUUUG-CGUAAAUUAUUUGGCGCAUUC--UCCCAGCCAUGUAGUUUGCACAGCAAUUAACUGCAAAACUUUAACCGCAAACUAAGCGUAAGGUAAACAAGUAAUCGA ...............(((-((((((((((.((((......--.....)))).)))))))))...(((......)))...((((((..(((.......)))))))))......)))).... ( -24.50) >DroSim_CAF1 20875 118 - 1 CAAAUUAAUUUCAAUUUGCCGUAAAUUAUUUGGCGCAUUC--UCCCAGCCAUGUAGUUUGCACAGCAAUUAACUGCAAAACUUUAACCGCAAACUAAGCGUAAGGUAAACAAGUAAUCGA ..............(((((((((((((((.((((......--.....)))).)))))))))...(((......)))...........(((.......)))...))))))........... ( -25.30) >DroEre_CAF1 24106 118 - 1 CAAAUUAAUUUCAAUUUGCCGUAAAUUAUUCGGCGCAUUC--UCACAGUCAUGUAGUUUGUACAGCAAUUAACUGCAAAACUUUAACCGCAAACUAAGCGUAAGGUAAACAAGUAAUCGA .................((((.........))))(((..(--(.(((....)))))..)))...(((......)))...((((((..(((.......))))))))).............. ( -18.60) >DroYak_CAF1 18483 120 - 1 CAAAUUAAUUUCAAUUUGCCGUAAAUUAUUCGGCGCAUUCCCUCACUGCCAUGCAGUUUGUGCAGCAAUUAACUGCAAAACUUUAACCGCAAACUAAGCGUAAGGUAAACAAGUAAUCGA .................((((.........))))((((......(((((...)))))..)))).(((......)))...((((((..(((.......))))))))).............. ( -22.10) >consensus CAAAUUAAUUUCAAUUUGCCGUAAAUUAUUUGGCGCAUUC__UCCCAGCCAUGUAGUUUGCACAGCAAUUAACUGCAAAACUUUAACCGCAAACUAAGCGUAAGGUAAACAAGUAAUCGA ..............(((((((((((((((.((((.............)))).)))))))))...(((......)))...........(((.......)))...))))))........... (-18.64 = -19.24 + 0.60)

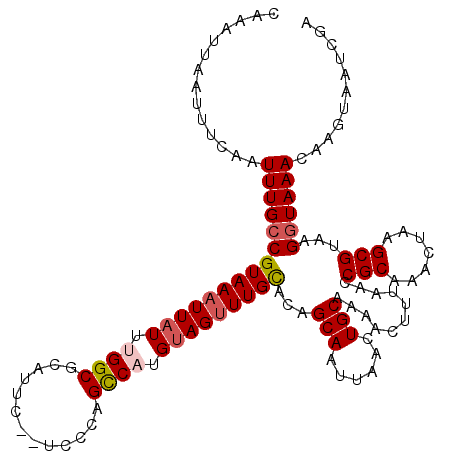
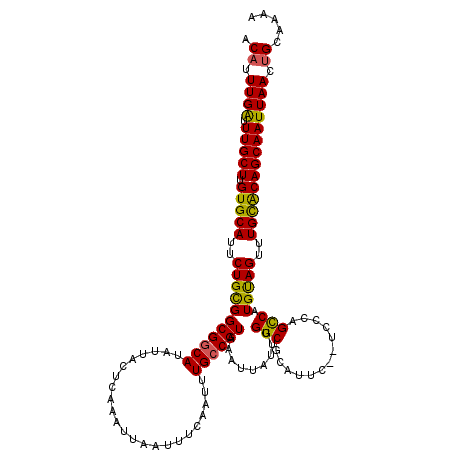
| Location | 532,753 – 532,871 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.26 |
| Mean single sequence MFE | -32.64 |
| Consensus MFE | -28.74 |
| Energy contribution | -27.94 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.20 |
| Mean z-score | -3.17 |
| Structure conservation index | 0.88 |
| SVM decision value | 4.40 |
| SVM RNA-class probability | 0.999889 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 532753 118 - 22224390 ACAUUUGAUUUGCUUGUGCAUUCUGCGGCGGCAUAUUACUCAAAUUAAUUUCAAUUUGCCGUAAAUUAUUUGGCGCAUUC--UCCCAGCCAUGUAGAUUGCACAGCAAUUAACUGCAAAA .((.((((((.(((.(((((.((((((((((((.(((...............))).))))))........((((......--.....)))))))))).)))))))))))))).))..... ( -35.46) >DroSec_CAF1 20758 117 - 1 ACACUUGAUUUGCUUGUGCAUUCUGCGGCGGCAUAUUACUCAAAUUAAUUUCAAUUUG-CGUAAAUUAUUUGGCGCAUUC--UCCCAGCCAUGUAGUUUGCACAGCAAUUAACUGCAAAA .((.((((((.(((.(((((..(((((..(((...((((.((((((......))))))-.))))......(((.(.....--.))))))).)))))..)))))))))))))).))..... ( -34.30) >DroSim_CAF1 20915 118 - 1 ACACUUGAUUUGCUUGUGCAUUCUGCGGCGGCAUAUUACUCAAAUUAAUUUCAAUUUGCCGUAAAUUAUUUGGCGCAUUC--UCCCAGCCAUGUAGUUUGCACAGCAAUUAACUGCAAAA .((.((((((.(((.(((((..(((((((((((.(((...............))).))))))........((((......--.....)))))))))..)))))))))))))).))..... ( -34.26) >DroEre_CAF1 24146 118 - 1 ACAUUUGGUUUGCUUGUGCAUUCUGUGGCGGCAUUUUACUCAAAUUAAUUUCAAUUUGCCGUAAAUUAUUCGGCGCAUUC--UCACAGUCAUGUAGUUUGUACAGCAAUUAACUGCAAAA .((.((((((.(((.(((((..(((((.(((...(((((.((((((......))))))..)))))....))).)))....--..(((....)))))..)))))))))))))).))..... ( -26.20) >DroYak_CAF1 18523 120 - 1 GCUUUUGGCUUGCUGGUGCAUUCUGUGGCGGCAUAUUACCCAAAUUAAUUUCAAUUUGCCGUAAAUUAUUCGGCGCAUUCCCUCACUGCCAUGCAGUUUGUGCAGCAAUUAACUGCAAAA ((..((((.(((((.(..((..(((((((((..........(((((......)))))((((.........))))...........)))))..))))..))..))))))))))..)).... ( -33.00) >consensus ACAUUUGAUUUGCUUGUGCAUUCUGCGGCGGCAUAUUACUCAAAUUAAUUUCAAUUUGCCGUAAAUUAUUUGGCGCAUUC__UCCCAGCCAUGUAGUUUGCACAGCAAUUAACUGCAAAA .((.((((.(((((.(((((..(((((((((((.......................)))))).........(((.............))).)))))..)))))))))))))).))..... (-28.74 = -27.94 + -0.80)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:36:57 2006