
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,414,391 – 4,414,544 |
| Length | 153 |
| Max. P | 0.998764 |

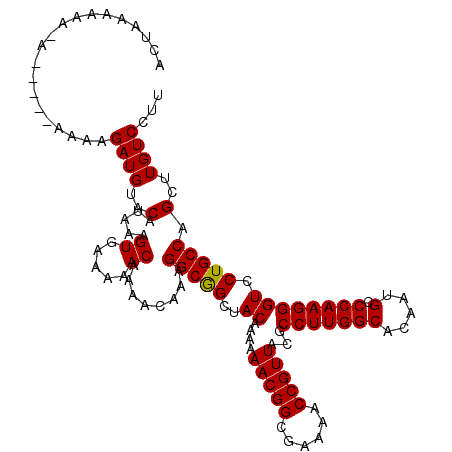
| Location | 4,414,391 – 4,414,504 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.26 |
| Mean single sequence MFE | -23.66 |
| Consensus MFE | -23.52 |
| Energy contribution | -23.36 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.99 |
| SVM decision value | 3.22 |
| SVM RNA-class probability | 0.998764 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4414391 113 - 22224390 ACUAAAAA--A-----AAAAGAUGUACAUAAGGUGAAAAACAAAACAAAGGCGGCUACAAAAAACGGCGAAAACCGUUACGCCUUGGCACAAUGCCCAAGGGUCCUGCCAGCUUGUCCUU ........--.-----...........(((((((.....))........(((((..((....(((((......)))))...(((((((.....).)))))))).)))))..))))).... ( -24.30) >DroSec_CAF1 16818 115 - 1 ACUUAAAAAAA-----AAAAGAUGUACAUAAAGUGAAAAACAAAACAAAGGCGGCUACAAAAAACGGCGAAAACCGUUACGCCUUGGCACAAUGCCCAAGGGUCCUGCCAGCUUGUCCUU ...........-----....((((..(.....((.....))........(((((..((....(((((......)))))...(((((((.....).)))))))).))))).)..))))... ( -23.30) >DroSim_CAF1 18289 120 - 1 ACUUAAAAAAAAAUAAAAAAGAUGUACAUAAAGUGAAAAACAAAACAAAGGCGGCUACAAAAAACGGCGAAAACCGUUACGCCUUGGCACAAUGCCCAAGGGUCCUGCCAGCUUGUCCUU ....................((((..(.....((.....))........(((((..((....(((((......)))))...(((((((.....).)))))))).))))).)..))))... ( -23.30) >DroEre_CAF1 17830 114 - 1 ACUAAAAAA-A-----AGAAGAUGUACAUAAAGUGAAAAACAAAACAAAGGCAGCUACAAAAAACGGCGAAAACCGUUACGCCUUGGCACAAUGCCCAAGGGUCCUGCCAGCUUGUCCUU .........-.-----....((((..(.....((.....))........(((((..((....(((((......)))))...(((((((.....).)))))))).))))).)..))))... ( -24.00) >DroYak_CAF1 17519 111 - 1 ACUAAAUAA-A-----A---GAUGUACAAAAAGUGAAAAACAAAACAAAGGCGGCUACAAAAAACGGCGAAAACCGUUACGCCUUGGCACAAUGCCCAAGGGUCCUGCCAGCUUGUCCUU .........-.-----.---...(.((((...((.....))........(((((..((....(((((......)))))...(((((((.....).)))))))).)))))...)))).).. ( -23.40) >consensus ACUAAAAAA_A_____AAAAGAUGUACAUAAAGUGAAAAACAAAACAAAGGCGGCUACAAAAAACGGCGAAAACCGUUACGCCUUGGCACAAUGCCCAAGGGUCCUGCCAGCUUGUCCUU ....................((((..(.....((.....))........(((((..((....(((((......)))))...(((((((.....).)))))))).))))).)..))))... (-23.52 = -23.36 + -0.16)


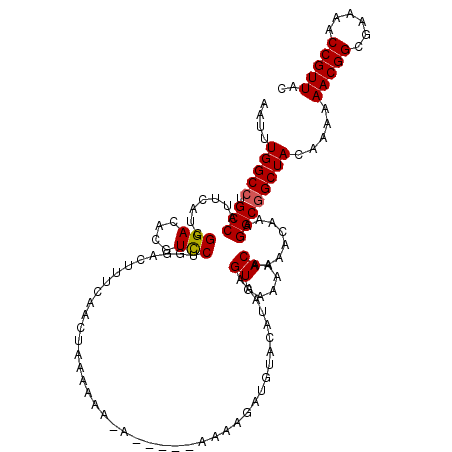
| Location | 4,414,431 – 4,414,544 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.29 |
| Mean single sequence MFE | -22.63 |
| Consensus MFE | -19.00 |
| Energy contribution | -19.04 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.71 |
| SVM RNA-class probability | 0.996498 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4414431 113 - 22224390 AAUUUGGCCUGCCAUUCAUCGGACACGUCCUGGACUUUCAACUAAAAA--A-----AAAAGAUGUACAUAAGGUGAAAAACAAAACAAAGGCGGCUACAAAAAACGGCGAAAACCGUUAC ....(((((.(((...(((((((....)))(((........)))....--.-----....))))........((.....))........)))))))).....(((((......))))).. ( -23.00) >DroSec_CAF1 16858 115 - 1 AAUUUGGCCUGCCAUUCAACGGACACGUCCUGGACUUUCAACUUAAAAAAA-----AAAAGAUGUACAUAAAGUGAAAAACAAAACAAAGGCGGCUACAAAAAACGGCGAAAACCGUUAC ....(((((.(((.((((..(((....))))))).(((((.(((.......-----..............))))))))...........)))))))).....(((((......))))).. ( -22.40) >DroSim_CAF1 18329 120 - 1 AAUUUGGCCUGCCAUUCAACGGACACGUUCUGGACUUUCAACUUAAAAAAAAAUAAAAAAGAUGUACAUAAAGUGAAAAACAAAACAAAGGCGGCUACAAAAAACGGCGAAAACCGUUAC ....(((((.(((.(((((((....)))..)))).(((((.(((..........................))))))))...........)))))))).....(((((......))))).. ( -21.47) >DroEre_CAF1 17870 113 - 1 AAUUUGGCCUGCCAUUCAUCGGACACGUCCAGGAC-UUCAACUAAAAAA-A-----AGAAGAUGUACAUAAAGUGAAAAACAAAACAAAGGCAGCUACAAAAAACGGCGAAAACCGUUAC ..((((..(((((...(((((((....)))((...-.....))......-.-----....))))........((.....))........)))))...)))).(((((......))))).. ( -23.00) >DroYak_CAF1 17559 111 - 1 AAUUUGGCCUGCCAUUCAUCGGACACGUCCAGGACAUUCAACUAAAUAA-A-----A---GAUGUACAAAAAGUGAAAAACAAAACAAAGGCGGCUACAAAAAACGGCGAAAACCGUUAC ....(((((.(((...(((((((....)))((.........))......-.-----.---))))........((.....))........)))))))).....(((((......))))).. ( -23.30) >consensus AAUUUGGCCUGCCAUUCAUCGGACACGUCCUGGACUUUCAACUAAAAAA_A_____AAAAGAUGUACAUAAAGUGAAAAACAAAACAAAGGCGGCUACAAAAAACGGCGAAAACCGUUAC ....(((((.(((.......(((....)))..........................................((.....))........)))))))).....(((((......))))).. (-19.00 = -19.04 + 0.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:10:33 2006