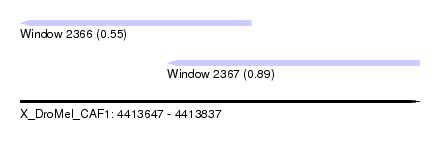
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,413,647 – 4,413,837 |
| Length | 190 |
| Max. P | 0.892493 |

| Location | 4,413,647 – 4,413,757 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.60 |
| Mean single sequence MFE | -32.26 |
| Consensus MFE | -25.60 |
| Energy contribution | -25.56 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.547105 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4413647 110 - 22224390 UUCCGGAUCAAAUUGUAAUACGCAUGUUGACCGCUCCGAAAACGGGAUGACCAAGAUCUCGGU---------GAACUGGUCAGCUAAAUGAAAUAUGAGUUCAAAGGGGCUUG-GAAUGG ((((((.((((..(((.....)))..))))))(((((.....((((((.......)))))).(---------(((((.((((......)))....).))))))..)))))..)-)))... ( -31.40) >DroSec_CAF1 16060 110 - 1 UUCCGGAUCAAAUUGUAAUACGCAUGUUGACCGCUCCGAAAACGGGAUGACCAAGAUCUCGGU---------GAACUGGUCAGCUAAAUGAAAUAUGAGUUCAAAGGGGCCAG-GAAUGG ((((((.((((..(((.....)))..))))))(((((.....((((((.......)))))).(---------(((((.((((......)))....).))))))..)))))..)-)))... ( -31.20) >DroSim_CAF1 17507 109 - 1 UUCCGGAUCAAAUUGUAAUACGCAUGUUGACCGCUCCGAAAACGGGAUGACCAAGAUCUCGGU---------GAACUGGUCAGCUAAAUGAAAUAUGAGUUCAAAG-GGCCAG-GAAUGG ((((((.((((..(((.....)))..))))))((.((.....((((((.......)))))).(---------(((((.((((......)))....).))))))..)-)))..)-)))... ( -27.10) >DroEre_CAF1 17081 120 - 1 UUCCGGAUCAAAUUGUAAUACGCAUGUUGACCGCUCCGAAAACGGGAUGACCAAGAUCUCGGUGGACUCGGUGGACUGGUCAGCCAAAUGAAAUAUGAGUUCAAAGGGGCUAGGGAAUGA ((((((.((((..(((.....)))..))))))(((((.....((((((.......)))))).((((((((.(((.((....))))).........))))))))..)))))...))))... ( -36.70) >DroYak_CAF1 16803 110 - 1 UUCCGGAUCAAAUUGUAAUACGCAUGUUGACCGCUCCGAAAACGGGAUGACCAAGAUCUCGGU---------GGACUGGUCAGCUAAAUGAAAUAUGAGUUCAAAGGGGCUCG-GAAUGA ((((...(((...(((.....))).((((((((.((((....((((((.......)))))).)---------))).))))))))....))).....((((((....)))))))-)))... ( -34.90) >consensus UUCCGGAUCAAAUUGUAAUACGCAUGUUGACCGCUCCGAAAACGGGAUGACCAAGAUCUCGGU_________GAACUGGUCAGCUAAAUGAAAUAUGAGUUCAAAGGGGCUAG_GAAUGG ((((((.((((..(((.....)))..))))))(((((.....((((((.......))))))...........(((((.((((......)))....).)))))...)))))..).)))... (-25.60 = -25.56 + -0.04)



| Location | 4,413,717 – 4,413,837 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.74 |
| Mean single sequence MFE | -26.79 |
| Consensus MFE | -24.58 |
| Energy contribution | -25.38 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.892493 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4413717 120 - 22224390 GGUCGCCAAAAGCCAGGAGCAGUAAGGAGAAGGGAAUAAAAGAAGUGACUGCGUCAUAAAACAAAUUCCUAAUCAAGGGAUUCCGGAUCAAAUUGUAAUACGCAUGUUGACCGCUCCGAA (((........))).(((((.....((((..((((((.......(((((...))))).......))))))..((....))))))((.((((..(((.....)))..)))))))))))... ( -30.04) >DroSec_CAF1 16130 116 - 1 GGUCGCCAAAAGCCAGGAGCAGUAAUGUGAAG----UAAAAAAAGUGACUGCGUCAUAAAACAAAUUCCUAAUCAAGGGAUUCCGGAUCAAAUUGUAAUACGCAUGUUGACCGCUCCGAA (((........))).(((((.((..(((((.(----((...........))).)))))..)).(((((((.....)))))))..((.((((..(((.....)))..)))))))))))... ( -27.90) >DroSim_CAF1 17576 116 - 1 GGUCGCCAAAAGCCAGGAGCAGUAAGGAGAAG----UAAAAAAAGUGACUGCGUCAUAAAACAAAUUCCUAAUCAAGGGAUUCCGGAUCAAAUUGUAAUACGCAUGUUGACCGCUCCGAA (((........))).(((((.....((((...----........(((((...)))))........(((((.....)))))))))((.((((..(((.....)))..)))))))))))... ( -29.40) >DroEre_CAF1 17161 115 - 1 GGUCGCCAAAAGCCAGAAGCAAUAAGGAGAAA----UAAA-AAAGUGACUGCGUCAUAAAACAAAUUCCUAAUCAAGGGAUUCCGGAUCAAAUUGUAAUACGCAUGUUGACCGCUCCGAA (((........)))...........((((...----....-...(((((...)))))......(((((((.....))))))).(((.((((..(((.....)))..)))))))))))... ( -23.30) >DroYak_CAF1 16873 115 - 1 GGUCGCCAAAAGCCAGAAGCAAUAAGGAGAAG----UAAA-AAAGUGACUGCGUCAUAAAACAAAUUCCUAAUCAAGGGAUUCCGGAUCAAAUUGUAAUACGCAUGUUGACCGCUCCGAA (((........)))...........((((...----....-...(((((...)))))......(((((((.....))))))).(((.((((..(((.....)))..)))))))))))... ( -23.30) >consensus GGUCGCCAAAAGCCAGGAGCAGUAAGGAGAAG____UAAAAAAAGUGACUGCGUCAUAAAACAAAUUCCUAAUCAAGGGAUUCCGGAUCAAAUUGUAAUACGCAUGUUGACCGCUCCGAA (((........))).(((((.....((((...............(((((...)))))........(((((.....)))))))))((.((((..(((.....)))..)))))))))))... (-24.58 = -25.38 + 0.80)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:10:31 2006