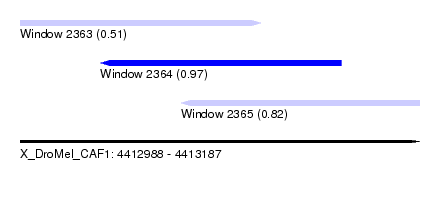
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,412,988 – 4,413,187 |
| Length | 199 |
| Max. P | 0.968563 |

| Location | 4,412,988 – 4,413,108 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.81 |
| Mean single sequence MFE | -26.42 |
| Consensus MFE | -23.60 |
| Energy contribution | -23.40 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.89 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.512061 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4412988 120 + 22224390 AACGGUCGAGGAAAGUUCCCAUCGAGCAAAUGAAAUUAAGUAAAUAAAUAAAUGGACGCGAGAGAUCGCGAGAAAAAUGCAAAUGAGCCUGCUCCGGCAAAAGGGACAUUAAGUAGCCGG ..(((..(...((.(((((......(((.......(((......))).........(((((....))))).......)))......(((......)))....))))).))...)..))). ( -25.20) >DroSec_CAF1 15422 119 + 1 AACGGUCGAGGAAAGUUCCCAUCGGGCAAAUGAAAUUAAGCAAAUAAAUAAAUGGACGCGAGAGAUCGCGAGAAAAAUGCAAAUGAGCCGUGUCCGGCAA-AGGGACAUUAAGUAGCCGG ..(((..(...((.(((((..(((.(((..((........))..............(((((....))))).......)))...)))((((....))))..-.))))).))...)..))). ( -29.00) >DroSim_CAF1 16434 119 + 1 AACGGUCGAGGAAAGUUCCCAUCGGGCAAAUGAAAUUAAGCAAAUAAAUAAAUGGACGCGAGAGAUCGCGAGAAAAAUGCAAAUGAGCCGGGUCCGGCAA-AGGGACAUUAAGUAGCCGG ..(((..(...((.(((((..(((.(((..((........))..............(((((....))))).......)))...)))((((....))))..-.))))).))...)..))). ( -30.90) >DroEre_CAF1 16455 119 + 1 AACGGUCGAGGAAAGUUCCCAGCGAGCAAAUGAAAUUAAACAAAUAAAUAAAUGGACGCGAGAGAUCGCGAGAAAAAUGCAAAUGAGCUUGGUCCGGCAA-AGGGACAUUAAGUAGCCGG ..(((..(...((.(((((..((.......((........))..........(((((((((....)))).........((......))...)))))))..-.))))).))...)..))). ( -25.20) >DroYak_CAF1 16169 119 + 1 AACGGUCGAGGAAAGUUCCCAGCGAGCAAAUGAAAUUAAACAAAUAAAUAAAUGGACGGGAGAGAUCGCGAGAAAAAUGCAAAUGAGCUUGGUCCGGCAA-AGGGACAUUAAGUAGCCGG ..(((..(...((.(((((..((.......((........))..........(((((.((.....((....)).....((......)))).)))))))..-.))))).))...)..))). ( -21.80) >consensus AACGGUCGAGGAAAGUUCCCAUCGAGCAAAUGAAAUUAAGCAAAUAAAUAAAUGGACGCGAGAGAUCGCGAGAAAAAUGCAAAUGAGCCUGGUCCGGCAA_AGGGACAUUAAGUAGCCGG ..(((..(...((.(((((......(((..((........))..............(((((....))))).......)))......(((......)))....))))).))...)..))). (-23.60 = -23.40 + -0.20)

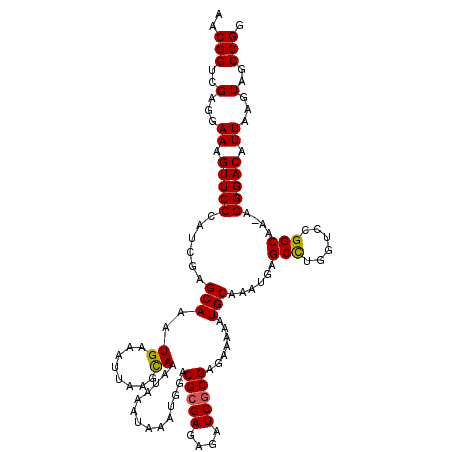
| Location | 4,413,028 – 4,413,148 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.64 |
| Mean single sequence MFE | -28.65 |
| Consensus MFE | -23.95 |
| Energy contribution | -23.75 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.968563 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4413028 120 - 22224390 CAACCCCUUUAUGGUCUUAAAACUGGACAAUGGUCAGCUACCGGCUACUUAAUGUCCCUUUUGCCGGAGCAGGCUCAUUUGCAUUUUUCUCGCGAUCUCUCGCGUCCAUUUAUUUAUUUA .....((.....)).........((((((((((((.(((.(((((.................)))))))).))).))))............((((....)))))))))............ ( -30.93) >DroSec_CAF1 15462 119 - 1 CAACCCCUGCUUGGUCUUAAAGCUGGACAAUGGUCAGCUACCGGCUACUUAAUGUCCCU-UUGCCGGACACGGCUCAUUUGCAUUUUUCUCGCGAUCUCUCGCGUCCAUUUAUUUAUUUG ............(((...((((..(((((..(((.(((.....))))))...)))))))-)))))((((...((......)).........((((....))))))))............. ( -28.80) >DroSim_CAF1 16474 119 - 1 CAACCCCUGCUCGGUCUUAAAACUGGACAAUGGUCAGCUACCGGCUACUUAAUGUCCCU-UUGCCGGACCCGGCUCAUUUGCAUUUUUCUCGCGAUCUCUCGCGUCCAUUUAUUUAUUUG ........(((.(((((...((..(((((..(((.(((.....))))))...)))))..-))...))))).)))................(((((....)))))................ ( -28.90) >DroEre_CAF1 16495 119 - 1 CAACCCCUUGCUGGUCCUAAAACUGGACAAUGGUCAGCUACCGGCUACUUAAUGUCCCU-UUGCCGGACCAAGCUCAUUUGCAUUUUUCUCGCGAUCUCUCGCGUCCAUUUAUUUAUUUG .........((((((((...((..(((((..(((.(((.....))))))...)))))..-))...))))).)))................(((((....)))))................ ( -30.20) >DroYak_CAF1 16209 119 - 1 CAACCCCUUCUUGGUCUUAAAACUGGACAAUGGUCAGCUACCGGCUACUUAAUGUCCCU-UUGCCGGACCAAGCUCAUUUGCAUUUUUCUCGCGAUCUCUCCCGUCCAUUUAUUUAUUUG .........((((((((...((..(((((..(((.(((.....))))))...)))))..-))...)))))))).....((((.........))))......................... ( -24.40) >consensus CAACCCCUUCUUGGUCUUAAAACUGGACAAUGGUCAGCUACCGGCUACUUAAUGUCCCU_UUGCCGGACCAGGCUCAUUUGCAUUUUUCUCGCGAUCUCUCGCGUCCAUUUAUUUAUUUG .............((((.......))))(((((.(((((.(((((.................)))))....))))................((((....))))).))))).......... (-23.95 = -23.75 + -0.20)


| Location | 4,413,068 – 4,413,187 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.68 |
| Mean single sequence MFE | -35.52 |
| Consensus MFE | -23.79 |
| Energy contribution | -23.79 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.819247 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4413068 119 - 22224390 AAUGAGACCAAAGACCAAAGACCGA-AGACGCACGAGCAGCAACCCCUUUAUGGUCUUAAAACUGGACAAUGGUCAGCUACCGGCUACUUAAUGUCCCUUUUGCCGGAGCAGGCUCAUUU ((((((.((...(((((((((((((-((..((.......)).....))))..))))))............))))).(((.(((((.................)))))))).)))))))). ( -34.73) >DroSec_CAF1 15502 110 - 1 AAUGAGACCAAAGAC-------CGA-AGAC-CACGAGCAGCAACCCCUGCUUGGUCUUAAAGCUGGACAAUGGUCAGCUACCGGCUACUUAAUGUCCCU-UUGCCGGACACGGCUCAUUU ((((((.((..((.(-------(((-((((-((..(((((......))))))))))))..(((((.((....)))))))..)))))......(((((..-.....))))).)))))))). ( -40.10) >DroSim_CAF1 16514 111 - 1 AAUGAGACCAAAGAC-------CGA-AGACGCACGAGCAGCAACCCCUGCUCGGUCUUAAAACUGGACAAUGGUCAGCUACCGGCUACUUAAUGUCCCU-UUGCCGGACCCGGCUCAUUU ((((((.((...(((-------((.-.......(((((((......)))))))((((.......))))..))))).....(((((..............-..)))))....)))))))). ( -39.59) >DroEre_CAF1 16535 119 - 1 AAUGAGACCAAAGACCAAAGACCCACAGACCCACGACCAACAACCCCUUGCUGGUCCUAAAACUGGACAAUGGUCAGCUACCGGCUACUUAAUGUCCCU-UUGCCGGACCAAGCUCAUUU ((((((......(((((........(((......(((((.(((....))).)))))......))).....))))).....(((((..............-..)))))......)))))). ( -29.91) >DroYak_CAF1 16249 119 - 1 AAUGAGACCAAAGACCAUAGACCCACAGACCCAGGAGCAACAACCCCUUCUUGGUCUUAAAACUGGACAAUGGUCAGCUACCGGCUACUUAAUGUCCCU-UUGCCGGACCAAGCUCAUUU ((((((......((((((.(.((...(((((.(((((.........))))).))))).......)).).)))))).....(((((..............-..)))))......)))))). ( -33.29) >consensus AAUGAGACCAAAGACCA_AGACCGA_AGACCCACGAGCAGCAACCCCUUCUUGGUCUUAAAACUGGACAAUGGUCAGCUACCGGCUACUUAAUGUCCCU_UUGCCGGACCAGGCUCAUUU ((((((((((.......................((((.((......)).))))((((.......))))..))))).(((.(((((.................)))))....)))))))). (-23.79 = -23.79 + -0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:10:29 2006