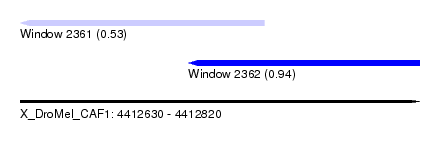
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,412,630 – 4,412,820 |
| Length | 190 |
| Max. P | 0.941264 |

| Location | 4,412,630 – 4,412,746 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.49 |
| Mean single sequence MFE | -22.44 |
| Consensus MFE | -12.68 |
| Energy contribution | -13.18 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.57 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.529189 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4412630 116 - 22224390 ACAUAUGUACA----UAUAGCAUAUACAUACAUAUAACACUCAUUUCGCGGCAAUCAUUCGGUACCUAACAGCUGUGAUCCGCGAUCCCAGAUCAAGUCACUAAUUAUAUGCCUCGAUUU ..(((((((..----((((...))))..)))))))..........(((.((((.....(..((..((.....(((.((((...)))).)))....))..))..).....)))).)))... ( -21.40) >DroSec_CAF1 15078 116 - 1 ACAUAUGUACAUGCAUAUAGCAUAUA----CAUAUAACACUCAUUUCGCGGCAAUCAUUCGGUACCUAACAGCUGUGAUCCGCGAUCCCAGAUCAAGUCACUAAUUAUAUGCCUCGAUUU ..(((((((.((((.....)))).))----)))))..........(((.((((.....(..((..((.....(((.((((...)))).)))....))..))..).....)))).)))... ( -27.00) >DroSim_CAF1 16090 116 - 1 ACAUAUGUACAUGCAUAUAGCAUAUA----CAUAUAACACUCAUUUCGCGGCAAUCAUUCGGUACCUAACAGCUGUGAUCCGCGAUCCCAGAUCAAGUCACUAAUUAUAUGCCUCGAUUU ..(((((((.((((.....)))).))----)))))..........(((.((((.....(..((..((.....(((.((((...)))).)))....))..))..).....)))).)))... ( -27.00) >DroEre_CAF1 16093 98 - 1 ACAUAUGUACAUAUAUAUAGCAU--------AUAUAACACUCAUUUCGCGGCAAUCAUUCGCUACCUAACAGCUGUGA--------------UCAAAUCACUAAUUAUAUGCCUCGAUUU ..((((((...........))))--------))............(((.((((.......(((.......))).((((--------------(...)))))........)))).)))... ( -16.60) >DroYak_CAF1 15826 107 - 1 ACAUAUGUAUAUACAUA-----U--------AUAUAACACUCAUUUUGCGGCAAUUAUUUGGUACAUAACAGCUGUGAUCCCCGAUCCCAGAUCAAGUCACUAAUUAUAUGCCUCGAUUU ...((((((((....))-----)--------))))).........(((.((((..((.(((((((.......(((.((((...)))).))).....)).))))).))..)))).)))... ( -20.20) >consensus ACAUAUGUACAUACAUAUAGCAUAUA____CAUAUAACACUCAUUUCGCGGCAAUCAUUCGGUACCUAACAGCUGUGAUCCGCGAUCCCAGAUCAAGUCACUAAUUAUAUGCCUCGAUUU ..((((((....))))))...........................(((.((((......((((........))))(((((..........)))))..............)))).)))... (-12.68 = -13.18 + 0.50)

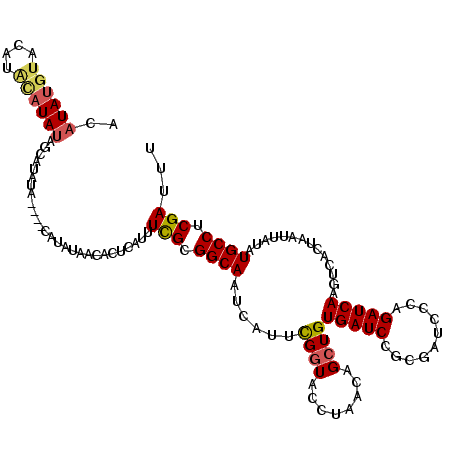

| Location | 4,412,710 – 4,412,820 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.02 |
| Mean single sequence MFE | -32.28 |
| Consensus MFE | -21.56 |
| Energy contribution | -21.48 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.941264 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4412710 110 - 22224390 ------GAAACCUAAGCCAAAGGGGCCAACAAAGCUCACUCGGGGGCAAACUCGUUUAUGUCCGCUUGGGUUCAAGUGAGACAUAUGUACA----UAUAGCAUAUACAUACAUAUAACAC ------(((.((((((((..((((((.......)))).))..)(((((..........)))))))))))))))..(((....(((((((..----((((...))))..)))))))..))) ( -27.40) >DroSec_CAF1 15158 110 - 1 ------GCAACCCAAGCCAAAGGGGCCAACAAAGCUCUAACGGGGGCAAACUCGUUUAUGUCCGCUUGGGUUCAAGUGAGACAUAUGUACAUGCAUAUAGCAUAUA----CAUAUAACAC ------(.(((((((((....(((((.......)))))((((((......)))))).......))))))))))..(((....(((((((.((((.....)))).))----)))))..))) ( -36.30) >DroSim_CAF1 16170 110 - 1 ------GCAACCCAAGCCAAAGGGGCCAACAAAGCUCUAUCGGGGGCAAACUCGUUUAUGUUCGCUUGGGUUCAAGUGAGACAUAUGUACAUGCAUAUAGCAUAUA----CAUAUAACAC ------(.(((((((((....(((((.......)))))..((((......)))).........))))))))))..(((....(((((((.((((.....)))).))----)))))..))) ( -34.10) >DroEre_CAF1 16159 112 - 1 AAUAGGAAACCCUAAGCCAAAGGGGCCAACAAAGCUCUCUCGGGGGCAAACUCGUUUAUGUCCGCUUGGGUUCAAGUGAGACAUAUGUACAUAUAUAUAGCAU--------AUAUAACAC ....((...((((.......)))).))......(((((....)))))......((.((((((((((((....)))))).))))))...)).(((((((...))--------))))).... ( -31.40) >DroYak_CAF1 15906 105 - 1 --UACGGAACCCUAAGCCGAAGGGGCCAACAAAGCUUUCUCGGGGACAAACUCGUUUAUGUCCGCUUGGGUUCAAGUGAGACAUAUGUAUAUACAUA-----U--------AUAUAACAC --....((((((.(((((((..((((.......))))..))).(((((..........)))))))))))))))..((.....((((((....)))))-----)--------.....)).. ( -32.20) >consensus ______GAAACCUAAGCCAAAGGGGCCAACAAAGCUCUCUCGGGGGCAAACUCGUUUAUGUCCGCUUGGGUUCAAGUGAGACAUAUGUACAUACAUAUAGCAUAUA____CAUAUAACAC ...............(((.....))).......(((((....)))))......((.((((((((((((....)))))).))))))...)).............................. (-21.56 = -21.48 + -0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:10:26 2006