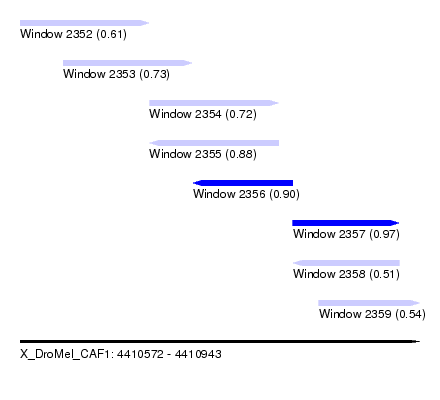
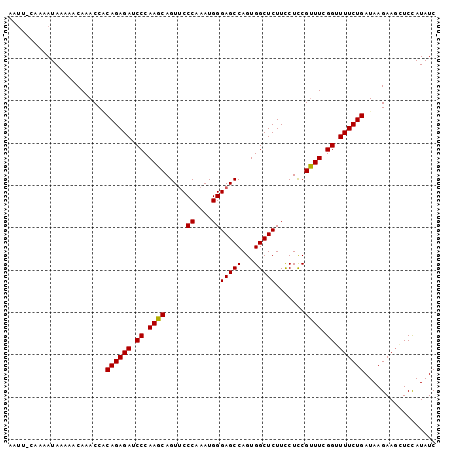
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,410,572 – 4,410,943 |
| Length | 371 |
| Max. P | 0.971856 |

| Location | 4,410,572 – 4,410,692 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.91 |
| Mean single sequence MFE | -25.19 |
| Consensus MFE | -24.33 |
| Energy contribution | -24.08 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.613134 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4410572 120 + 22224390 UUCCAUUCCCAUUCCCAUUCCCAUUCCAAAUUCCCACUUCCACUUCCACUUCCAUUUCCACCUGGGGCGCACAACUGUCUUGCUCUUGGGCGGCUGUCUUCGGUCCGCUGAGCAAAUGGC ...................................................((((((.(.((.((((((.((....))..)))))).))(((((((....))).))))...).)))))). ( -23.50) >DroSec_CAF1 13104 114 + 1 UUUCAUUCUCA------UUCCCAUUCCAAGUUCCCACUUCCACUUCCAUUUCCAUUUCCACCUGGGGCGCACAACUGUCUUGCUCUUGGGCGGCUGUCUUCGGUCCGCUGAGCAAAUGGC ...........------...(((((....(((((((..........................))))).))..........(((((...((((((((....))).))))))))))))))). ( -24.37) >DroEre_CAF1 13950 107 + 1 UUCCAUUCCCA------UUCCCAUUCC-------CAUUGCCACUUCCAUUCCCAUUUCCACCUGGGGCGCACAACUGUCUUGCUCUUGGGCGGCUGUCUUCGGUCCGCUGAGCAAAUGGC ...........------..........-------....((((.......(((((........)))))............((((((...((((((((....))).))))))))))).)))) ( -27.40) >DroYak_CAF1 13254 101 + 1 UUCCAUUUCGA------UUCCCAUUCC-------------CACUUACAUUCCCAUUUCCACCUGGGGCGCACAACUGUCUUGCUCUUGGGCGGCUGUCUUCGGUCCGCUGAGCAAAUGGC ...........------...(((((..-------------.........(((((........))))).............(((((...((((((((....))).))))))))))))))). ( -25.50) >consensus UUCCAUUCCCA______UUCCCAUUCC_______CACUUCCACUUCCAUUCCCAUUUCCACCUGGGGCGCACAACUGUCUUGCUCUUGGGCGGCUGUCUUCGGUCCGCUGAGCAAAUGGC ....................(((((........................(((((........))))).............(((((...((((((((....))).))))))))))))))). (-24.33 = -24.08 + -0.25)


| Location | 4,410,612 – 4,410,732 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.19 |
| Mean single sequence MFE | -40.88 |
| Consensus MFE | -39.80 |
| Energy contribution | -40.05 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.730324 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4410612 120 + 22224390 CACUUCCACUUCCAUUUCCACCUGGGGCGCACAACUGUCUUGCUCUUGGGCGGCUGUCUUCGGUCCGCUGAGCAAAUGGCGGGAAAGCAAUCCCAACCAAGUCCGGCCCAUAUGGCAAAA ..............((((((..((((.((.((....(((((((((...((((((((....))).)))))))))))..)))((((......))))......)).)).))))..))).))). ( -41.50) >DroSec_CAF1 13138 120 + 1 CACUUCCAUUUCCAUUUCCACCUGGGGCGCACAACUGUCUUGCUCUUGGGCGGCUGUCUUCGGUCCGCUGAGCAAAUGGCGGGAAAGCAAUCCCAACCAAGUCCGGCCCAUAUGGCAAAA ..............((((((..((((.((.((....(((((((((...((((((((....))).)))))))))))..)))((((......))))......)).)).))))..))).))). ( -41.50) >DroEre_CAF1 13977 120 + 1 CACUUCCAUUCCCAUUUCCACCUGGGGCGCACAACUGUCUUGCUCUUGGGCGGCUGUCUUCGGUCCGCUGAGCAAAUGGCGGGAAAGCAAUCCCAACCAAGUCCGGCCCAUAUGGCAAAA ..............((((((..((((.((.((....(((((((((...((((((((....))).)))))))))))..)))((((......))))......)).)).))))..))).))). ( -41.50) >DroYak_CAF1 13275 120 + 1 CACUUACAUUCCCAUUUCCACCUGGGGCGCACAACUGUCUUGCUCUUGGGCGGCUGUCUUCGGUCCGCUGAGCAAAUGGCGGGAAAGCAAUCCAAACCAAGUCCGGCCCAUAUGGCAAAA ..............((((((..((((.((.((.......((((((...((((((((....))).))))))))))).(((..(((......)))...))).)).)).))))..))).))). ( -39.00) >consensus CACUUCCAUUCCCAUUUCCACCUGGGGCGCACAACUGUCUUGCUCUUGGGCGGCUGUCUUCGGUCCGCUGAGCAAAUGGCGGGAAAGCAAUCCCAACCAAGUCCGGCCCAUAUGGCAAAA ..............((((((..((((.((.((....(((((((((...((((((((....))).)))))))))))..)))((((......))))......)).)).))))..))).))). (-39.80 = -40.05 + 0.25)



| Location | 4,410,692 – 4,410,812 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -37.65 |
| Consensus MFE | -33.30 |
| Energy contribution | -33.30 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.722648 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4410692 120 + 22224390 GGGAAAGCAAUCCCAACCAAGUCCGGCCCAUAUGGCAAAAGAUAUGGAGCUUCUUAUCAGAAACCCAAAACGGAGAAAGAGCCACUGGCUCCCAUUUGGGAACUGCUUGGGAUCUCUGGG ((((......))))..(((((((((((((((((........)))))).)))......(((...((((((..((((..((.....))..))))..))))))..)))....))).)).))). ( -37.90) >DroSec_CAF1 13218 120 + 1 GGGAAAGCAAUCCCAACCAAGUCCGGCCCAUAUGGCAAAAGAUAUGGAGCUUCUUAUCAGAAAACCGAAACGGAGGAAGAGCCACUGGCUCCCAUUUGGGAACUGCUUGGGAUCUCUGCG ......((((((((((.((..((((((((((((........)))))).)))......((((...(((...))).((..(((((...))))))).)))))))..)).)))))))...))). ( -38.50) >DroEre_CAF1 14057 120 + 1 GGGAAAGCAAUCCCAACCAAGUCCGGCCCAUAUGGCAAAAGAUAUAGACGUUCUUAUCAGAAAACCGAAGCGAAGGAAGAGCCACUGGCUCCCAUUUGGGUACUGCUUGGGAUCUCUGUG ((((......))))..((((((...(((((.((((.....((((.((.....))))))......((........))..(((((...))))))))).)))))...)))))).......... ( -38.30) >DroYak_CAF1 13355 120 + 1 GGGAAAGCAAUCCAAACCAAGUCCGGCCCAUAUGGCAAAAGAUAUAGACCUUCUUAUCAGAAGACCGAGGCGGAGAAAGAGCCACUGGCUCCCAUUUGGGAACUGCUUGGGAUCUUUGGG .(((......)))...((.......(((.....))).((((((......(((((....))))).((.((((((.......(((...)))((((....)))).)))))).)))))))))). ( -35.90) >consensus GGGAAAGCAAUCCCAACCAAGUCCGGCCCAUAUGGCAAAAGAUAUAGACCUUCUUAUCAGAAAACCGAAACGGAGAAAGAGCCACUGGCUCCCAUUUGGGAACUGCUUGGGAUCUCUGGG ((((......))))..((((((....((((.((((.....((((.((.....))))))....................(((((...))))))))).))))....)))))).......... (-33.30 = -33.30 + 0.00)



| Location | 4,410,692 – 4,410,812 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -39.25 |
| Consensus MFE | -36.53 |
| Energy contribution | -37.15 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.876376 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4410692 120 - 22224390 CCCAGAGAUCCCAAGCAGUUCCCAAAUGGGAGCCAGUGGCUCUUUCUCCGUUUUGGGUUUCUGAUAAGAAGCUCCAUAUCUUUUGCCAUAUGGGCCGGACUUGGUUGGGAUUGCUUUCCC ...((..(((((((.(((((((((.(((((((((...)))))...........(((((((((....)))))).))).........)))).))))...))).)).)))))))..))..... ( -43.20) >DroSec_CAF1 13218 120 - 1 CGCAGAGAUCCCAAGCAGUUCCCAAAUGGGAGCCAGUGGCUCUUCCUCCGUUUCGGUUUUCUGAUAAGAAGCUCCAUAUCUUUUGCCAUAUGGGCCGGACUUGGUUGGGAUUGCUUUCCC .(((...(((((((.(((.(((.(((((((((((...))))).....)))))).((((((((....)))))..((((((........)))))))))))).))).))))))))))...... ( -39.40) >DroEre_CAF1 14057 120 - 1 CACAGAGAUCCCAAGCAGUACCCAAAUGGGAGCCAGUGGCUCUUCCUUCGCUUCGGUUUUCUGAUAAGAACGUCUAUAUCUUUUGCCAUAUGGGCCGGACUUGGUUGGGAUUGCUUUCCC ...((..(((((((.((((.((((.(((((((((...)))))....(((...((((....))))...)))...............)))).))))....)).)).)))))))..))..... ( -37.50) >DroYak_CAF1 13355 120 - 1 CCCAAAGAUCCCAAGCAGUUCCCAAAUGGGAGCCAGUGGCUCUUUCUCCGCCUCGGUCUUCUGAUAAGAAGGUCUAUAUCUUUUGCCAUAUGGGCCGGACUUGGUUUGGAUUGCUUUCCC .(((((....((((((....((((.(((((((((...)))))............(..(((((....)))))..)...........)))).))))...).))))))))))........... ( -36.90) >consensus CCCAGAGAUCCCAAGCAGUUCCCAAAUGGGAGCCAGUGGCUCUUCCUCCGCUUCGGUUUUCUGAUAAGAAGCUCCAUAUCUUUUGCCAUAUGGGCCGGACUUGGUUGGGAUUGCUUUCCC ......((((((((.(((((((((.(((((((((...)))))............((((((((....))))))))...........)))).))))...))).)).))))))))........ (-36.53 = -37.15 + 0.62)


| Location | 4,410,732 – 4,410,825 |
|---|---|
| Length | 93 |
| Sequences | 3 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 87.46 |
| Mean single sequence MFE | -23.37 |
| Consensus MFE | -21.32 |
| Energy contribution | -21.10 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.900427 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4410732 93 - 22224390 --------AAUAAAAACAAACCCCAGAGAUCCCAAGCAGUUCCCAAAUGGGAGCCAGUGGCUCUUUCUCCGUUUUGGGUUUCUGAUAAGAAGCUCCAUAUC --------...............(((((((((.((((.((((((....))))))....((........)))))).)))))))))................. ( -25.60) >DroSec_CAF1 13258 101 - 1 AAUUCCAAAAUAAAAACAAACCGCAGAGAUCCCAAGCAGUUCCCAAAUGGGAGCCAGUGGCUCUUCCUCCGUUUCGGUUUUCUGAUAAGAAGCUCCAUAUC .......................((((((.((.((((.((((((....))))))....((........)))))).)).))))))................. ( -21.60) >DroEre_CAF1 14097 101 - 1 AAUUACAAAAUAAAAACAAACCACAGAGAUCCCAAGCAGUACCCAAAUGGGAGCCAGUGGCUCUUCCUUCGCUUCGGUUUUCUGAUAAGAACGUCUAUAUC .......................((((((.((.((((.....((....))(((((...))))).......)))).)).))))))................. ( -22.90) >consensus AAUU_CAAAAUAAAAACAAACCACAGAGAUCCCAAGCAGUUCCCAAAUGGGAGCCAGUGGCUCUUCCUCCGUUUCGGUUUUCUGAUAAGAAGCUCCAUAUC .......................((((((.((.((((.....((....))(((((...))))).......)))).)).))))))................. (-21.32 = -21.10 + -0.22)

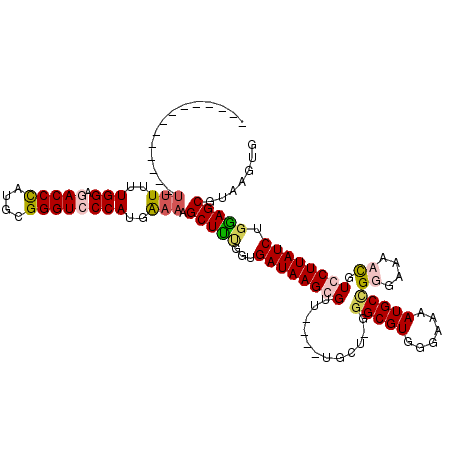
| Location | 4,410,825 – 4,410,924 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.32 |
| Mean single sequence MFE | -38.08 |
| Consensus MFE | -25.99 |
| Energy contribution | -27.17 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.971856 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4410825 99 + 22224390 ----------------UUUUGGAUACCUAUGCGGGUCCCAUGGAAAGCUCGGGUGAUAAGCGUU----UGCU-GCGCGUGGGAAAAUGCUGGGAAAACGUCCUUAUCUGGAGCGUAAGUG ----------------.........((((.(((..(((((((...(((..(.((.....)).).----.)))-...)))))))...)))))))...((((((......))).)))..... ( -30.70) >DroSec_CAF1 13359 102 + 1 --------------UUUUUUGGAUACCUAUGCGGGUCCCAUGGAAAGCUUUGGUGAUAAGCGUU----UGCUUGGGCGUGGGAAAAUGCUGGGAAAAUGUCCUUAUCUGGAGCGUAAGUG --------------......(((((((((.(((..(((((((..((((..((.(....).))..----.))))...)))))))...)))))))....))))).........((....)). ( -29.80) >DroEre_CAF1 14198 114 + 1 -----UUUUUUUGCUUUUUUGGAGACCCAUGCGGGUCCCAUGAAAAGCUUCGAUGAUAAGCGUUGCGUUGCU-GGGCGUGGGAAAAUGCCGGGAAACCUUGCUUAUCUGGAGCGCAAGUG -----....(((((((((.(((.(((((....)))))))).)))).((((((..((((((((..(.(((.((-.(((((......))))).)).)))).))))))))))))))))))).. ( -47.00) >DroYak_CAF1 13995 115 + 1 GUUCUCUCUUGUAUUUUUUUGGAGACCCAUGCGGGUCCCAUGAAAAGCUUUGGUGAUAAGCGUU----UUCU-GGGCGUGGGAAAAUGCCGGGAAACCUUGCUUAUCUGAAGCGUAAGCG (((..(.......(((((.(((.(((((....)))))))).)))))(((((...((((((((.(----((((-.(((((......))))).)))))...)))))))).))))))..))). ( -44.80) >consensus ______________UUUUUUGGAGACCCAUGCGGGUCCCAUGAAAAGCUUUGGUGAUAAGCGUU____UGCU_GGGCGUGGGAAAAUGCCGGGAAAACGUCCUUAUCUGGAGCGUAAGUG ..............(((..(((.(((((....))))))))..))).(((((...((((((((............(((((......)))))((....)).)))))))).)))))....... (-25.99 = -27.17 + 1.19)

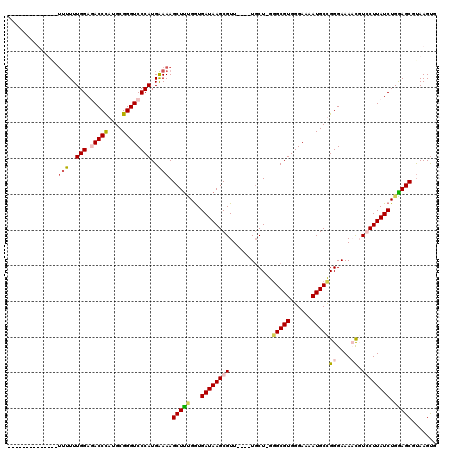
| Location | 4,410,825 – 4,410,924 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.32 |
| Mean single sequence MFE | -25.27 |
| Consensus MFE | -14.77 |
| Energy contribution | -15.65 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.58 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.514312 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4410825 99 - 22224390 CACUUACGCUCCAGAUAAGGACGUUUUCCCAGCAUUUUCCCACGCGC-AGCA----AACGCUUAUCACCCGAGCUUUCCAUGGGACCCGCAUAGGUAUCCAAAA---------------- .......((((..((((((..(((((.(...((..........))..-.).)----))))))))))....))))......(((((((......))).))))...---------------- ( -20.30) >DroSec_CAF1 13359 102 - 1 CACUUACGCUCCAGAUAAGGACAUUUUCCCAGCAUUUUCCCACGCCCAAGCA----AACGCUUAUCACCAAAGCUUUCCAUGGGACCCGCAUAGGUAUCCAAAAAA-------------- .(((((.(((((......))).....(((((............((....)).----...((((.......))))......)))))...)).)))))..........-------------- ( -18.40) >DroEre_CAF1 14198 114 - 1 CACUUGCGCUCCAGAUAAGCAAGGUUUCCCGGCAUUUUCCCACGCCC-AGCAACGCAACGCUUAUCAUCGAAGCUUUUCAUGGGACCCGCAUGGGUCUCCAAAAAAGCAAAAAAA----- .............(((((((..((....))(((..........))).-...........)))))))......((((((..((((((((....))))).))).)))))).......----- ( -32.00) >DroYak_CAF1 13995 115 - 1 CGCUUACGCUUCAGAUAAGCAAGGUUUCCCGGCAUUUUCCCACGCCC-AGAA----AACGCUUAUCACCAAAGCUUUUCAUGGGACCCGCAUGGGUCUCCAAAAAAAUACAAGAGAGAAC ..(((.(((((..(((((((....((((..(((..........))).-.)))----)..)))))))....))))......((((((((....))))).)))...........).)))... ( -30.40) >consensus CACUUACGCUCCAGAUAAGCAAGGUUUCCCAGCAUUUUCCCACGCCC_AGCA____AACGCUUAUCACCAAAGCUUUCCAUGGGACCCGCAUAGGUAUCCAAAAAA______________ ..(((((......).))))...((...((((((....(((((.((....))........((((.......))))......)))))...)).))))...)).................... (-14.77 = -15.65 + 0.88)

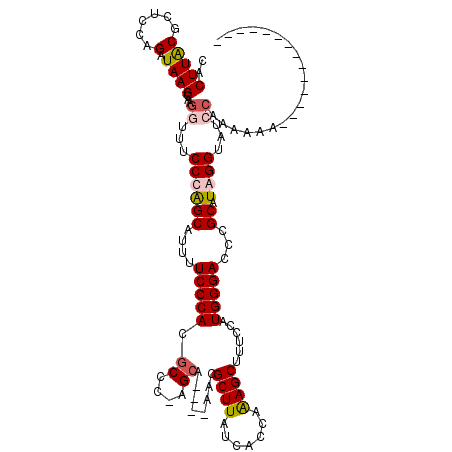
| Location | 4,410,849 – 4,410,943 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 88.95 |
| Mean single sequence MFE | -29.07 |
| Consensus MFE | -20.00 |
| Energy contribution | -21.25 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.542771 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4410849 94 + 22224390 UGGAAAGCUCGGGUGAUAAGCGUU----UGCU-GCGCGUGGGAAAAUGCUGGGAAAACGUCCUUAUCUGGAGCGUAAGUGUUGUCAUUUGAAUCCACAU ((((..((((((((((...(((((----(.((-..((((......))))..)).))))))..)))))).)))).((((((....))))))..))))... ( -27.30) >DroSec_CAF1 13385 95 + 1 UGGAAAGCUUUGGUGAUAAGCGUU----UGCUUGGGCGUGGGAAAAUGCUGGGAAAAUGUCCUUAUCUGGAGCGUAAGUGUUGUCAUUUGAAUCCACAU ((((..((((..(......(((((----(..((.(((((......))))).)).))))))......)..)))).((((((....))))))..))))... ( -25.90) >DroEre_CAF1 14233 98 + 1 UGAAAAGCUUCGAUGAUAAGCGUUGCGUUGCU-GGGCGUGGGAAAAUGCCGGGAAACCUUGCUUAUCUGGAGCGCAAGUGUUGUCAUUUGAAUCCACAU ......((((((..((((((((..(.(((.((-.(((((......))))).)).)))).)))))))))))))).((((((....))))))......... ( -33.30) >DroYak_CAF1 14035 94 + 1 UGAAAAGCUUUGGUGAUAAGCGUU----UUCU-GGGCGUGGGAAAAUGCCGGGAAACCUUGCUUAUCUGAAGCGUAAGCGUUGUCAUUUGAAUCCACAU ..........(((.((((((((.(----((((-.(((((......))))).)))))...))))))))(((.((....))....))).......)))... ( -29.80) >consensus UGAAAAGCUUUGGUGAUAAGCGUU____UGCU_GGGCGUGGGAAAAUGCCGGGAAAACGUCCUUAUCUGGAGCGUAAGUGUUGUCAUUUGAAUCCACAU ((((..(((((...((((((((............(((((......)))))((....)).)))))))).))))).((((((....))))))..))))... (-20.00 = -21.25 + 1.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:10:24 2006