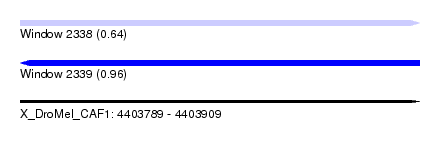
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,403,789 – 4,403,909 |
| Length | 120 |
| Max. P | 0.963702 |

| Location | 4,403,789 – 4,403,909 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.92 |
| Mean single sequence MFE | -45.37 |
| Consensus MFE | -40.32 |
| Energy contribution | -40.04 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.642303 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4403789 120 + 22224390 GCCCGCUGCCUUGGCUUUGCCCCUGCAAAGCCAGUCCACUGCUGCCCAUCUGAAUCAGGGAGUCAGUGUGGUCGCCAAUGCAGCCAGUCAUGCCAAUGUGGCCGAGGAUGCUGGCAUCGU (((.((..((((((((((((....))))))))....(((((((.(((..........))))).))))).((((((....(((........)))....))))))))))..)).)))..... ( -48.10) >DroSec_CAF1 6381 120 + 1 GCCCGCUGCCUUGGCUUUGCCCCUGCAAAGCCAGUCCACUGCCGCCCAUCUGAAUCAGGCAGUCAGUGUGGUCGCCAAUGCCGCCAGUCAUGCCCAUGUGGCGGAAGAUGCUGGCAUCGU ((((((((..((((((((((....))))))))))...((((((..............))))))))))).))).((((...((((((..(((....))))))))).......))))..... ( -48.14) >DroSim_CAF1 6473 120 + 1 GCCCGCUGCCUUGGCUUUGCCCCUGCAAAGCCAGUCCACUGCCGCCCAUGUGAAUCAGGGAGUCAGUGUGGUCGCCAAUGCCGCCAGUCAUGCCAAUGUGGCCGAAGAUGCUGGCAUCGU ((..((.(((((((((((((....))))))))))..(((((.(.(((.((.....))))).).))))).))).))....)).(((((.((((((.....))).....))))))))..... ( -46.20) >DroEre_CAF1 7093 120 + 1 GCCCGCUGCCUUGGCUUUGCCACUGCAAAGUCAAUCCACUGCUUCCCAUUUGAAUCAGGGAGUCAGUGUGGUCGCCAAUGCCGCCAGUCACGCCAAUGUGGCCGAAGAUGCAGGCAUCGU ...((.((((((((((((((....)))))))))((((((((.(((((..........))))).)))))((((.((....)).))))((((((....))))))....)))..))))).)). ( -43.60) >DroYak_CAF1 6603 120 + 1 CCCCGCUGCCUUGGCUUUGCCCCUGCAAAGUCAAUCCACUGCUGCCCAUCUUAAUCAGGGAGUCAGUGUGGUCGCCAAUGCCGCCAGUCAUGCCAAUGUGGCCGAAGAUGCUGGUAUCGU ....((.(((((((((((((....))))))))))..(((((((.(((..........))))).))))).))).))..(((..(((((.((((((.....))).....))))))))..))) ( -40.80) >consensus GCCCGCUGCCUUGGCUUUGCCCCUGCAAAGCCAGUCCACUGCUGCCCAUCUGAAUCAGGGAGUCAGUGUGGUCGCCAAUGCCGCCAGUCAUGCCAAUGUGGCCGAAGAUGCUGGCAUCGU ....((.(((((((((((((....))))))))))..(((((.(.(((..........))).).))))).))).)).......(((((.((((((.....))).....))))))))..... (-40.32 = -40.04 + -0.28)


| Location | 4,403,789 – 4,403,909 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.92 |
| Mean single sequence MFE | -48.89 |
| Consensus MFE | -44.36 |
| Energy contribution | -45.32 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.963702 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4403789 120 - 22224390 ACGAUGCCAGCAUCCUCGGCCACAUUGGCAUGACUGGCUGCAUUGGCGACCACACUGACUCCCUGAUUCAGAUGGGCAGCAGUGGACUGGCUUUGCAGGGGCAAAGCCAAGGCAGCGGGC .((((((.(((....((((((.....))).)))...)))))))))((..((.(((((.(((((..........))).)))))))...(((((((((....))))))))).))..)).... ( -50.80) >DroSec_CAF1 6381 120 - 1 ACGAUGCCAGCAUCUUCCGCCACAUGGGCAUGACUGGCGGCAUUGGCGACCACACUGACUGCCUGAUUCAGAUGGGCGGCAGUGGACUGGCUUUGCAGGGGCAAAGCCAAGGCAGCGGGC .(((((((.((.......(((.....))).......)))))))))((..((.(((((.((((((.........)))))))))))...(((((((((....))))))))).))..)).... ( -54.14) >DroSim_CAF1 6473 120 - 1 ACGAUGCCAGCAUCUUCGGCCACAUUGGCAUGACUGGCGGCAUUGGCGACCACACUGACUCCCUGAUUCACAUGGGCGGCAGUGGACUGGCUUUGCAGGGGCAAAGCCAAGGCAGCGGGC .(((((((.((....((((((.....))).)))...)))))))))((..((.(((((.(.(((((.....)).))).).)))))...(((((((((....))))))))).))..)).... ( -52.00) >DroEre_CAF1 7093 120 - 1 ACGAUGCCUGCAUCUUCGGCCACAUUGGCGUGACUGGCGGCAUUGGCGACCACACUGACUCCCUGAUUCAAAUGGGAAGCAGUGGAUUGACUUUGCAGUGGCAAAGCCAAGGCAGCGGGC .(((((((.((....((((((.....))).)))...)))))))))((..((.(((((..((((..........))))..)))))..(((.((((((....)))))).)))))..)).... ( -47.80) >DroYak_CAF1 6603 120 - 1 ACGAUACCAGCAUCUUCGGCCACAUUGGCAUGACUGGCGGCAUUGGCGACCACACUGACUCCCUGAUUAAGAUGGGCAGCAGUGGAUUGACUUUGCAGGGGCAAAGCCAAGGCAGCGGGG .((((.((.((....((((((.....))).)))...)))).))))((..((.(((((.(((((..........))).)))))))..(((.((((((....)))))).)))))..)).... ( -39.70) >consensus ACGAUGCCAGCAUCUUCGGCCACAUUGGCAUGACUGGCGGCAUUGGCGACCACACUGACUCCCUGAUUCAGAUGGGCAGCAGUGGACUGGCUUUGCAGGGGCAAAGCCAAGGCAGCGGGC .(((((((.((....((((((.....))).)))...)))))))))((..((.(((((.(((((..........))).)))))))...(((((((((....))))))))).))..)).... (-44.36 = -45.32 + 0.96)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:10:06 2006