
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,402,918 – 4,403,072 |
| Length | 154 |
| Max. P | 0.824168 |

| Location | 4,402,918 – 4,403,038 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.17 |
| Mean single sequence MFE | -30.94 |
| Consensus MFE | -27.40 |
| Energy contribution | -27.96 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.824168 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4402918 120 + 22224390 ACCCACAAAUGGCCCUUUUGGCAGAAACUUUCAUCCGCAUCGAUUCGGGGUUAUUAAUGAUCGGUCGAUCCAUUGACCCCGACAGAAAGUCAAAGAAGAAAUAGUAAUAAAACGGGUUCU ((((.......(((.....)))....(((((((((......)))(((((((((.....((((....))))...)))))))))..)))))).......................))))... ( -32.40) >DroSec_CAF1 5528 120 + 1 ACCCACAAAUGGCCCUUUUGGCAGAAACUUUCAUCCACAUCGAUGCGGGGUUAUUAAUGAUCGGUCGAUCCAUUGACCCCGACAGAAAGCCAAAGAAGAAAUAGUAAUCAAACGGGUUCU ..........((((((((((((.........((((......))))((((((((.....((((....))))...)))))))).......)))))))........((......))))))).. ( -33.10) >DroSim_CAF1 5611 120 + 1 ACCCACAAAUGGCCCUUUUGGCAGAAACUUUCAUCCGCAUCGAUGCGGGGUUAUUAAUGAUCGGUCGAUCCAUUGAACCCGACAGAAAGCCAAAGAAGAAAUAGUAAUCAAACGGGUUCU ..........((((((((((((.............((((....))))((((..(((((((((....))).))))))))))........)))))))........((......))))))).. ( -30.20) >DroEre_CAF1 6277 120 + 1 ACCCACAAAGGGCCCUUUUGGCAGAAACUUUCAUCCGCAUCGAUGCGGGGUUAUUAAUGAUCGGCCGAUCCAUUGACCCCGACAGAAAGUCAAAGAAGAAAUAGUCAUAAAACGGGUUCC .........(((((((((((...((...((((.((.(((....)))(((((((.....((((....))))...)))))))(((.....)))...)).))))...)).))))).)))))). ( -33.50) >DroYak_CAF1 5749 120 + 1 ACACACAAUGGGCCCUUUUGGCAGAAACUUUCAUCCGCAUCGAUGCUUGGUUAUUAAUGAUCGGUCGAUCCAUUGACCCCGACAGAAAGUCAAAGAAGAAAUAGUAAUAAAACGGGUUCU .........(((((((((((((...((((..((((......))))...))))..........((((((....))))))..........)))))))........((......)))))))). ( -25.50) >consensus ACCCACAAAUGGCCCUUUUGGCAGAAACUUUCAUCCGCAUCGAUGCGGGGUUAUUAAUGAUCGGUCGAUCCAUUGACCCCGACAGAAAGUCAAAGAAGAAAUAGUAAUAAAACGGGUUCU ..........((((((((((((.........((((......))))((((((((.....((((....))))...)))))))).......)))))))........((......))))))).. (-27.40 = -27.96 + 0.56)

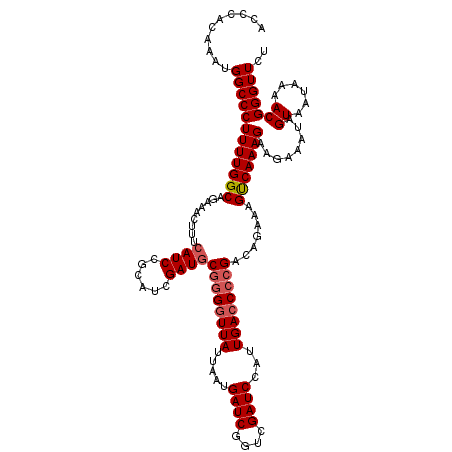

| Location | 4,402,958 – 4,403,072 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.05 |
| Mean single sequence MFE | -31.56 |
| Consensus MFE | -23.04 |
| Energy contribution | -23.84 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.560589 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4402958 114 + 22224390 CGAUUCGGGGUUAUUAAUGAUCGGUCGAUCCAUUGACCCCGACAGAAAGUCAAAGAAGAAAUAGUAAUAAAACGGGUUCUCGGAAGAAGAAAGCCCAACGAUGCUAGCCAAGCU------ ((.((((((((((.....((((....))))...)))))))(((.....)))...)))................((((((((....).)))..))))..))..(((.....))).------ ( -31.20) >DroSec_CAF1 5568 114 + 1 CGAUGCGGGGUUAUUAAUGAUCGGUCGAUCCAUUGACCCCGACAGAAAGCCAAAGAAGAAAUAGUAAUCAAACGGGUUCUCGGAAGAAGAAAGCCCAACGAUGCUGGCCAAGCU------ ...((((((((((.....((((....))))...)))))))).))....((((...........((......))((((((((....).)))..))))........))))......------ ( -32.90) >DroSim_CAF1 5651 114 + 1 CGAUGCGGGGUUAUUAAUGAUCGGUCGAUCCAUUGAACCCGACAGAAAGCCAAAGAAGAAAUAGUAAUCAAACGGGUUCUCGGAAGAAGAAAGCCCAACGAUGCUGGCCAAGCU------ ....((.((((..(((((((((....))).))))))))))........((((...........((......))((((((((....).)))..))))........))))...)).------ ( -28.30) >DroEre_CAF1 6317 116 + 1 CGAUGCGGGGUUAUUAAUGAUCGGCCGAUCCAUUGACCCCGACAGAAAGUCAAAGAAGAAAUAGUCAUAAAACGGGUUCCCGGAA---GAAAGCCCAAAGAUGCUAGCCAAGCUGG-GAU .(((..(((((((.....((((....))))...)))))))(((.....)))............))).......(((((..(....---)..)))))....((.(((((...)))))-.)) ( -34.40) >DroYak_CAF1 5789 117 + 1 CGAUGCUUGGUUAUUAAUGAUCGGUCGAUCCAUUGACCCCGACAGAAAGUCAAAGAAGAAAUAGUAAUAAAACGGGUUCUCCAAA---GAAAGCCCAAUGGUGCUAGCCAAGCUGGAGAU ....(((((((((.........((((((....))))))(((((.....)))......................(((((.((....---)).)))))...))...)))))))))....... ( -31.00) >consensus CGAUGCGGGGUUAUUAAUGAUCGGUCGAUCCAUUGACCCCGACAGAAAGUCAAAGAAGAAAUAGUAAUAAAACGGGUUCUCGGAAGAAGAAAGCCCAACGAUGCUAGCCAAGCU______ .....((((((((.....((((....))))...))))))))................................(((((.((.......)).)))))......(((.....)))....... (-23.04 = -23.84 + 0.80)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:10:03 2006