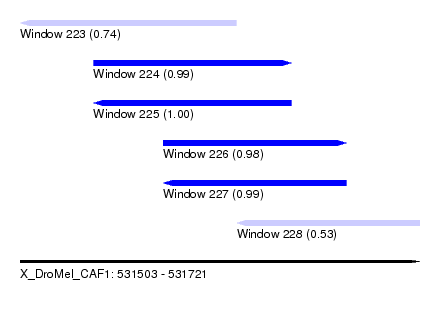
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 531,503 – 531,721 |
| Length | 218 |
| Max. P | 0.999561 |

| Location | 531,503 – 531,621 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.51 |
| Mean single sequence MFE | -18.48 |
| Consensus MFE | -16.01 |
| Energy contribution | -16.01 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.735585 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 531503 118 - 22224390 UCAUUUUAUUGGUGUAAUUUUUAAUUGCAUAUCAGCAAUUUAUAAAUAUUUACAUCGCAGCUGCAC--ACGCACACUCUCCCUUCUAUCAGGCCAAAUAAAUCAAUAACUGUCAAAAGCA ..........(((((((.(((.((((((......))))))...)))...)))))))((((.(((..--..))).......(((......)))................))))........ ( -17.20) >DroSec_CAF1 19784 116 - 1 UCAUUUUAUUGGUGUAAUUUUUAAUUGCAUAUCAGCAAUUUAUAAAUAUUUACAUCGCAGCUGCAC--AUGCACACUCUC--UCCUAUCGGGCCAAAUAAAUCAAUAACUGUCAGAAGCA ..........(((((((.(((.((((((......))))))...)))...)))))))((.(.(((..--..))))......--..((..(((.................)))..))..)). ( -17.93) >DroSim_CAF1 19587 116 - 1 UCAUUUUAUUGGUGUAAUUUUUAAUUGCAUAUCAGCAAUUUAUAAAUAUUUACAUCGCAGCUGCAC--ACGCACACUCUC--UUCUAUCGGGCCAAAUAAAUCAAUAACUGUCAGAAGCA ..........(((((((.(((.((((((......))))))...)))...)))))))((.(.(((..--..))))......--((((..(((.................)))..)))))). ( -19.93) >DroEre_CAF1 23183 119 - 1 UCAUUUUAUUGGUGUAAUUUUUAAUUGCAUAUCAGCAAUUUAUAAAUAUUUACAUCGCAGCUGCACACACGCACGCACUC-CUUCUAUAUGGCCAAAUAAAUCAAUAACUGUCAAAAGCA ..........(((((((.(((.((((((......))))))...)))...)))))))((.(((((......))).))....-........((((.................))))...)). ( -20.33) >DroYak_CAF1 17515 119 - 1 UCAUUUUAUUGGUGUAAUUUUUAAUUGCAUAUCAGCAAUUUAUAAAUAUUUACAUCGCAGCUGCCCACACGCACACACUC-UCGCUCUAUGGCCAAAUAAAUCAAUAACUGUCAAAAGCA ..........(((((((.(((.((((((......))))))...)))...)))))))((((.(((......))).......-..(((....)))...............))))........ ( -17.00) >consensus UCAUUUUAUUGGUGUAAUUUUUAAUUGCAUAUCAGCAAUUUAUAAAUAUUUACAUCGCAGCUGCAC__ACGCACACUCUC__UUCUAUCGGGCCAAAUAAAUCAAUAACUGUCAAAAGCA ..........(((((((.(((.((((((......))))))...)))...)))))))((.(.(((......))))................(((.................)))....)). (-16.01 = -16.01 + -0.00)

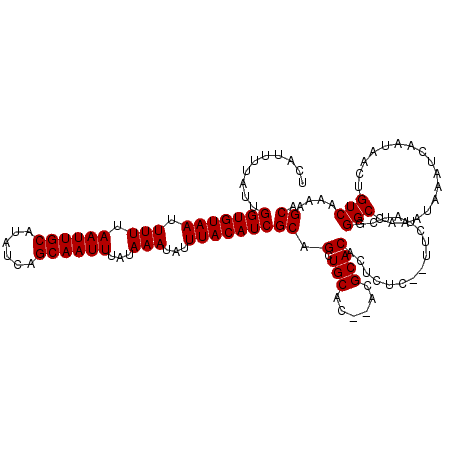
| Location | 531,543 – 531,651 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.94 |
| Mean single sequence MFE | -35.50 |
| Consensus MFE | -27.44 |
| Energy contribution | -27.48 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.90 |
| Structure conservation index | 0.77 |
| SVM decision value | 2.43 |
| SVM RNA-class probability | 0.993903 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 531543 108 + 22224390 GAGAGUGUGCGU--GUGCAGCUGCGAUGUAAAUAUUUAUAAAUUGCUGAUAUGCAAUUAAAAAUUACACCAAUAAAAUGACAGCUGCCUCAGGUGCAGCACCACAUGCAC---------- ......((((((--(((((((((((.(((((....)))))((((((......))))))...................)).)))))))....((((...))))))))))))---------- ( -32.90) >DroSec_CAF1 19822 107 + 1 GAGAGUGUGCAU--GUGCAGCUGCGAUGUAAAUAUUUAUAAAUUGCUGAUAUGCAAUUAAAAAUUACACCAAUAAAAUGACAGCUGC-UCAGGUGCAGCACCACAUGCAC---------- ......((((((--(((((((((((.(((((....)))))((((((......))))))...................)).)))))))-...((((...))))))))))))---------- ( -33.70) >DroSim_CAF1 19625 107 + 1 GAGAGUGUGCGU--GUGCAGCUGCGAUGUAAAUAUUUAUAAAUUGCUGAUAUGCAAUUAAAAAUUACACCAAUAAAAUGACAGCUGC-UCAGGUGCAGCACCACAUGCAC---------- ......((((((--(((((((((((.(((((....)))))((((((......))))))...................)).)))))))-...((((...))))))))))))---------- ( -33.30) >DroEre_CAF1 23222 110 + 1 GAGUGCGUGCGUGUGUGCAGCUGCGAUGUAAAUAUUUAUAAAUUGCUGAUAUGCAAUUAAAAAUUACACCAAUAAAAUGACAGCUGCCUCAGGUGCAGCUCCACAUGCAC---------- ......(((((((((.(.(((((((.(((((...((((...(((((......)))))))))..))))).)...............(((...)))))))))))))))))))---------- ( -32.70) >DroYak_CAF1 17554 120 + 1 GAGUGUGUGCGUGUGGGCAGCUGCGAUGUAAAUAUUUAUAAAUUGCUGAUAUGCAAUUAAAAAUUACACCAAUAAAAUGACAGCUGCCUCAGGUGCAGCACCACAUGCACAUGCAGAUGC ..(((((((((((((((((((((((.(((((....)))))((((((......))))))...................)).)))))))))..((((...))))))))))))))))...... ( -44.90) >consensus GAGAGUGUGCGU__GUGCAGCUGCGAUGUAAAUAUUUAUAAAUUGCUGAUAUGCAAUUAAAAAUUACACCAAUAAAAUGACAGCUGCCUCAGGUGCAGCACCACAUGCAC__________ ......((((((....(((((((((.(((((....)))))((((((......))))))...................)).)))))))....((((...))))..)))))).......... (-27.44 = -27.48 + 0.04)

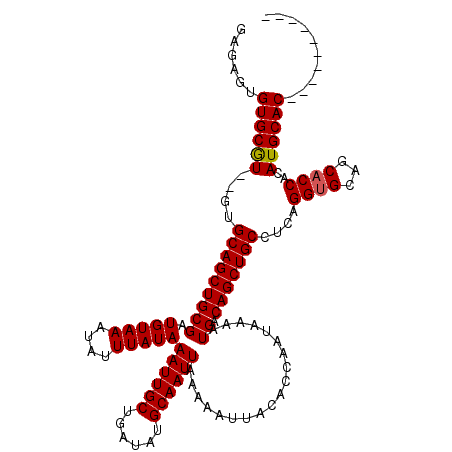
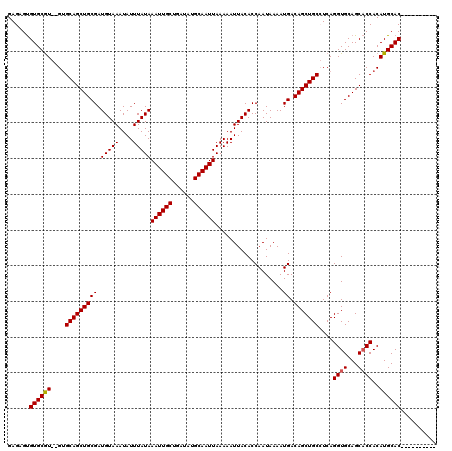
| Location | 531,543 – 531,651 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.94 |
| Mean single sequence MFE | -38.30 |
| Consensus MFE | -33.00 |
| Energy contribution | -33.20 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -4.97 |
| Structure conservation index | 0.86 |
| SVM decision value | 3.72 |
| SVM RNA-class probability | 0.999561 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 531543 108 - 22224390 ----------GUGCAUGUGGUGCUGCACCUGAGGCAGCUGUCAUUUUAUUGGUGUAAUUUUUAAUUGCAUAUCAGCAAUUUAUAAAUAUUUACAUCGCAGCUGCAC--ACGCACACUCUC ----------((((.((((..(((((......(((....)))........(((((((.(((.((((((......))))))...)))...))))))))))))..)))--).))))...... ( -36.00) >DroSec_CAF1 19822 107 - 1 ----------GUGCAUGUGGUGCUGCACCUGA-GCAGCUGUCAUUUUAUUGGUGUAAUUUUUAAUUGCAUAUCAGCAAUUUAUAAAUAUUUACAUCGCAGCUGCAC--AUGCACACUCUC ----------((((((((((((...))))...-((((((((.........(((((((.(((.((((((......))))))...)))...)))))))))))))))))--))))))...... ( -40.50) >DroSim_CAF1 19625 107 - 1 ----------GUGCAUGUGGUGCUGCACCUGA-GCAGCUGUCAUUUUAUUGGUGUAAUUUUUAAUUGCAUAUCAGCAAUUUAUAAAUAUUUACAUCGCAGCUGCAC--ACGCACACUCUC ----------((((.(((((((...))))...-((((((((.........(((((((.(((.((((((......))))))...)))...)))))))))))))))))--).))))...... ( -36.20) >DroEre_CAF1 23222 110 - 1 ----------GUGCAUGUGGAGCUGCACCUGAGGCAGCUGUCAUUUUAUUGGUGUAAUUUUUAAUUGCAUAUCAGCAAUUUAUAAAUAUUUACAUCGCAGCUGCACACACGCACGCACUC ----------((((.((((.((((((......(((....)))........(((((((.(((.((((((......))))))...)))...))))))))))))).))))...))))...... ( -38.00) >DroYak_CAF1 17554 120 - 1 GCAUCUGCAUGUGCAUGUGGUGCUGCACCUGAGGCAGCUGUCAUUUUAUUGGUGUAAUUUUUAAUUGCAUAUCAGCAAUUUAUAAAUAUUUACAUCGCAGCUGCCCACACGCACACACUC ((....)).(((((.(((((((...)))).(.(((((((((.........(((((((.(((.((((((......))))))...)))...)))))))))))))))))))).)))))..... ( -40.80) >consensus __________GUGCAUGUGGUGCUGCACCUGAGGCAGCUGUCAUUUUAUUGGUGUAAUUUUUAAUUGCAUAUCAGCAAUUUAUAAAUAUUUACAUCGCAGCUGCAC__ACGCACACUCUC ..........((((....((((...))))....((((((((.........(((((((.(((.((((((......))))))...)))...)))))))))))))))......))))...... (-33.00 = -33.20 + 0.20)

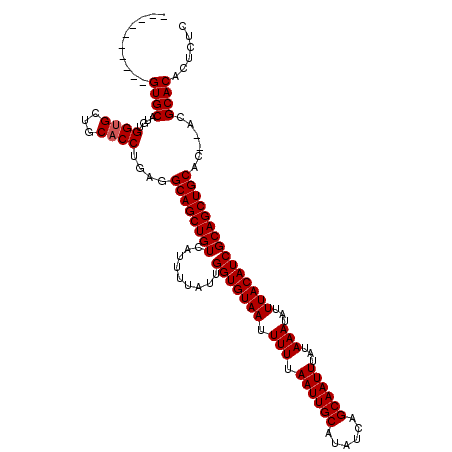
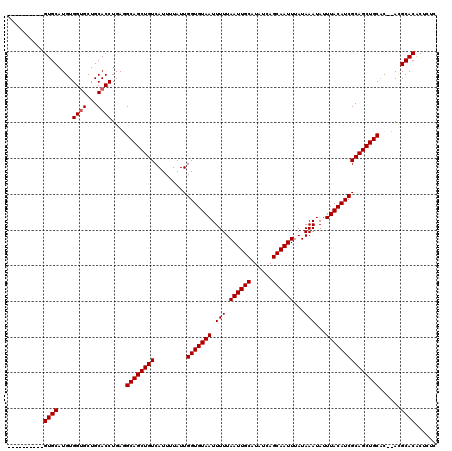
| Location | 531,581 – 531,681 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.04 |
| Mean single sequence MFE | -26.76 |
| Consensus MFE | -19.08 |
| Energy contribution | -19.88 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.38 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.979009 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 531581 100 + 22224390 AAUUGCUGAUAUGCAAUUAAAAAUUACACCAAUAAAAUGACAGCUGCCUCAGGUGCAGCACCACAUGCAC--------------------GCACAUGCAAAUUAAGCCCCGAUGCAGCUC ((((((......)))))).......................((((((.((.((.((.(((.....)))..--------------------((....)).......)))).)).)))))). ( -26.10) >DroSec_CAF1 19860 99 + 1 AAUUGCUGAUAUGCAAUUAAAAAUUACACCAAUAAAAUGACAGCUGC-UCAGGUGCAGCACCACAUGCAC--------------------GCACAUGCAAAUUAAGCCCCGAUGCAGCUC ((((((......)))))).......................((((((-((.((.((.(((.....)))..--------------------((....)).......)))).)).)))))). ( -25.50) >DroSim_CAF1 19663 99 + 1 AAUUGCUGAUAUGCAAUUAAAAAUUACACCAAUAAAAUGACAGCUGC-UCAGGUGCAGCACCACAUGCAC--------------------GCACAUGCAAAUUAAGCCCCGAUGCAGCUC ((((((......)))))).......................((((((-((.((.((.(((.....)))..--------------------((....)).......)))).)).)))))). ( -25.50) >DroEre_CAF1 23262 96 + 1 AAUUGCUGAUAUGCAAUUAAAAAUUACACCAAUAAAAUGACAGCUGCCUCAGGUGCAGCUCCACAUGCAC------------------------AUGCAAAUGAAGCACCAAUGCAGCUC ((((((......)))))).......................((((((....(((((.((.......)).(------------------------((....)))..)))))...)))))). ( -25.80) >DroYak_CAF1 17594 120 + 1 AAUUGCUGAUAUGCAAUUAAAAAUUACACCAAUAAAAUGACAGCUGCCUCAGGUGCAGCACCACAUGCACAUGCAGAUGCAGAUGCAGAUGCACAUGCAAAUGAAGCGCCAAUGCAUCUC ((((((......))))))........................((((((....).)))))......((((..(((....)))..))))((((((..(((.......)))....)))))).. ( -30.90) >consensus AAUUGCUGAUAUGCAAUUAAAAAUUACACCAAUAAAAUGACAGCUGCCUCAGGUGCAGCACCACAUGCAC____________________GCACAUGCAAAUUAAGCCCCGAUGCAGCUC ((((((......)))))).......................((((((....((.((.(((.....)))......................((....)).......)).))...)))))). (-19.08 = -19.88 + 0.80)

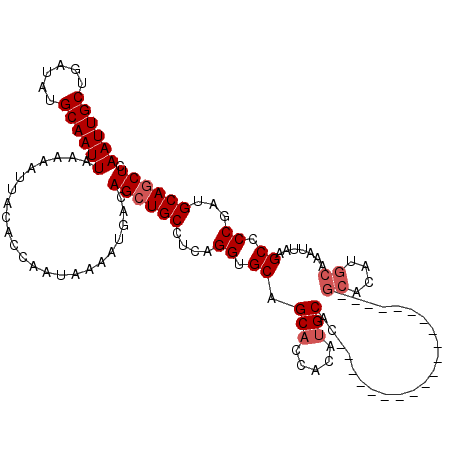
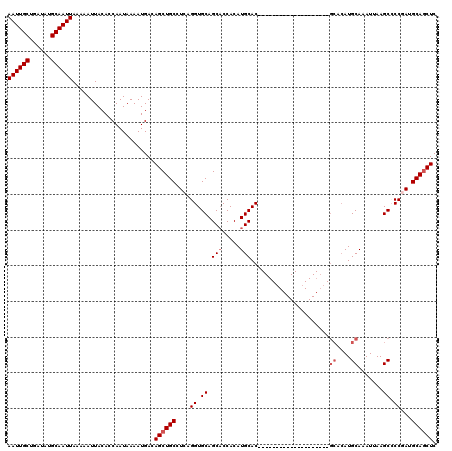
| Location | 531,581 – 531,681 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.04 |
| Mean single sequence MFE | -34.08 |
| Consensus MFE | -26.62 |
| Energy contribution | -29.58 |
| Covariance contribution | 2.96 |
| Combinations/Pair | 0.97 |
| Mean z-score | -2.97 |
| Structure conservation index | 0.78 |
| SVM decision value | 2.42 |
| SVM RNA-class probability | 0.993685 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 531581 100 - 22224390 GAGCUGCAUCGGGGCUUAAUUUGCAUGUGC--------------------GUGCAUGUGGUGCUGCACCUGAGGCAGCUGUCAUUUUAUUGGUGUAAUUUUUAAUUGCAUAUCAGCAAUU .((((((.(((((((.....(..(((((..--------------------..)))))..)....)).))))).))))))............(((((((.....))))))).......... ( -33.60) >DroSec_CAF1 19860 99 - 1 GAGCUGCAUCGGGGCUUAAUUUGCAUGUGC--------------------GUGCAUGUGGUGCUGCACCUGA-GCAGCUGUCAUUUUAUUGGUGUAAUUUUUAAUUGCAUAUCAGCAAUU .((((((.(((((((.....(..(((((..--------------------..)))))..)....)).)))))-))))))............(((((((.....))))))).......... ( -33.00) >DroSim_CAF1 19663 99 - 1 GAGCUGCAUCGGGGCUUAAUUUGCAUGUGC--------------------GUGCAUGUGGUGCUGCACCUGA-GCAGCUGUCAUUUUAUUGGUGUAAUUUUUAAUUGCAUAUCAGCAAUU .((((((.(((((((.....(..(((((..--------------------..)))))..)....)).)))))-))))))............(((((((.....))))))).......... ( -33.00) >DroEre_CAF1 23262 96 - 1 GAGCUGCAUUGGUGCUUCAUUUGCAU------------------------GUGCAUGUGGAGCUGCACCUGAGGCAGCUGUCAUUUUAUUGGUGUAAUUUUUAAUUGCAUAUCAGCAAUU .((((((...((((((((((.(((..------------------------..))).)))))...)))))....))))))............(((((((.....))))))).......... ( -30.70) >DroYak_CAF1 17594 120 - 1 GAGAUGCAUUGGCGCUUCAUUUGCAUGUGCAUCUGCAUCUGCAUCUGCAUGUGCAUGUGGUGCUGCACCUGAGGCAGCUGUCAUUUUAUUGGUGUAAUUUUUAAUUGCAUAUCAGCAAUU ((.(((((..((((((.((..((((((((((..(((....)))..)))))))))))).))))))(((((.(((((....))).)).....)))))..........))))).))....... ( -40.10) >consensus GAGCUGCAUCGGGGCUUAAUUUGCAUGUGC____________________GUGCAUGUGGUGCUGCACCUGAGGCAGCUGUCAUUUUAUUGGUGUAAUUUUUAAUUGCAUAUCAGCAAUU .((((((.(((((((.....(..((((((((((((((......))))))))))))))..)....)).))))).))))))............(((((((.....))))))).......... (-26.62 = -29.58 + 2.96)

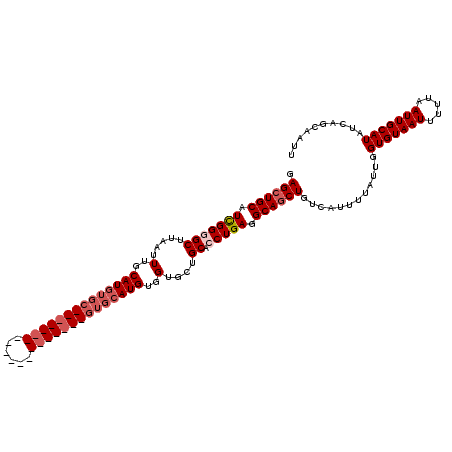
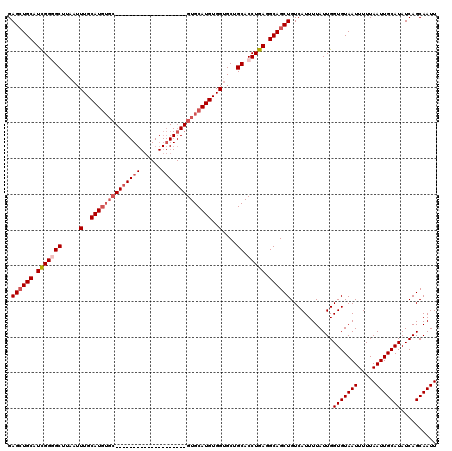
| Location | 531,621 – 531,721 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.15 |
| Mean single sequence MFE | -32.46 |
| Consensus MFE | -23.52 |
| Energy contribution | -26.48 |
| Covariance contribution | 2.96 |
| Combinations/Pair | 0.97 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.72 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.532276 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 531621 100 - 22224390 UGAAAAUCUCGAACUUUUCCGCACACAUUUGCAUUUUAAAGAGCUGCAUCGGGGCUUAAUUUGCAUGUGC--------------------GUGCAUGUGGUGCUGCACCUGAGGCAGCUG .(((((........))))).(((......))).........((((((.(((((((.....(..(((((..--------------------..)))))..)....)).))))).)))))). ( -32.20) >DroSec_CAF1 19900 99 - 1 UGAAAAUCUCGAACUUUCGCGCACACAGUUGCAUUUUAAAGAGCUGCAUCGGGGCUUAAUUUGCAUGUGC--------------------GUGCAUGUGGUGCUGCACCUGA-GCAGCUG (((((....(((....))).(((......))).)))))...((((((.(((((((.....(..(((((..--------------------..)))))..)....)).)))))-)))))). ( -31.40) >DroSim_CAF1 19703 99 - 1 UGAAAAUCUCGAACUUUCGCGCACACAGUUGCAUUUUAAAGAGCUGCAUCGGGGCUUAAUUUGCAUGUGC--------------------GUGCAUGUGGUGCUGCACCUGA-GCAGCUG (((((....(((....))).(((......))).)))))...((((((.(((((((.....(..(((((..--------------------..)))))..)....)).)))))-)))))). ( -31.40) >DroEre_CAF1 23302 96 - 1 UGAAAAUCUCGAACCCUUUCGCCCACAUUUGCAUUUUAAAGAGCUGCAUUGGUGCUUCAUUUGCAU------------------------GUGCAUGUGGAGCUGCACCUGAGGCAGCUG (((((.....(((....)))((........)).)))))...((((((...((((((((((.(((..------------------------..))).)))))...)))))....)))))). ( -27.30) >DroYak_CAF1 17634 119 - 1 -GAAAAUCUCGAACCCUUUCGCCCACAUUUGCAUUUUAAAGAGAUGCAUUGGCGCUUCAUUUGCAUGUGCAUCUGCAUCUGCAUCUGCAUGUGCAUGUGGUGCUGCACCUGAGGCAGCUG -...................(((((....((((((((...)))))))).))).)).((((.((((((((((..(((....)))..)))))))))).)))).(((((.......))))).. ( -40.00) >consensus UGAAAAUCUCGAACUUUUGCGCACACAUUUGCAUUUUAAAGAGCUGCAUCGGGGCUUAAUUUGCAUGUGC____________________GUGCAUGUGGUGCUGCACCUGAGGCAGCUG ....................((........)).........((((((.(((((((.....(..((((((((((((((......))))))))))))))..)....)).))))).)))))). (-23.52 = -26.48 + 2.96)

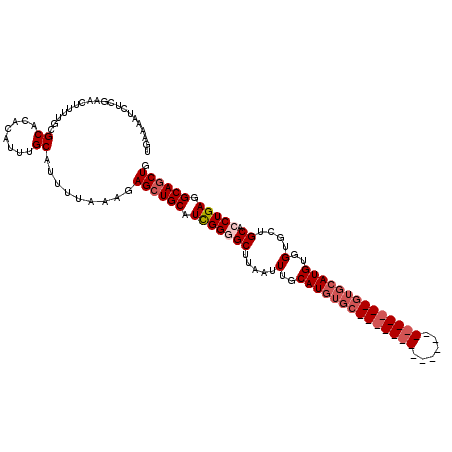
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:36:50 2006