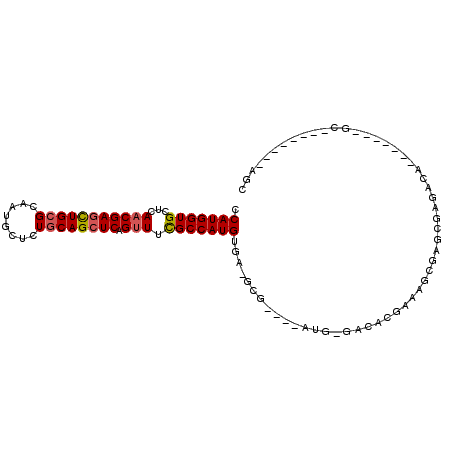
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,381,841 – 4,381,933 |
| Length | 92 |
| Max. P | 0.871440 |

| Location | 4,381,841 – 4,381,933 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 78.34 |
| Mean single sequence MFE | -29.69 |
| Consensus MFE | -18.68 |
| Energy contribution | -18.90 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.63 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.516750 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4381841 92 + 22224390 GCU--------GCUCGCUUUCGUCUUGUUCGCUUUCGUGUC-CAU----CGC-UCACAUGGCGAAACUGAGCUGCAGAGCAUUGCGCAGCUCGUUGAGCACCAUGG (.(--------((((..(((((((.(((..((.........-...----.))-..))).)))))))..(((((((..........)))))))...))))).).... ( -29.72) >DroSec_CAF1 167007 76 + 1 CCU--------GC-------UGUCUCGCUCGGUUU--------------CGC-UCACAUGGCGAAACUGAGCUGCAGAGCAUUGCGCAGCUCGUUGAGCACCAUGG (((--------(.-------((.((((((((((((--------------(((-(.....)))))))))))))....((((........))))...))))).)).)) ( -33.20) >DroSim_CAF1 154502 76 + 1 CCU--------GC-------UGUCUCGCUCGGUUU--------------CGC-UCACAUGGCGAAACUGAGCUGCAGAGCAUUGCGCAGCUCGUUGAGCACCAUGG (((--------(.-------((.((((((((((((--------------(((-(.....)))))))))))))....((((........))))...))))).)).)) ( -33.20) >DroEre_CAF1 177850 85 + 1 GCU--------GC-------UGUCUCGCUUGCAUUCGUGUC-CAU----CGC-UCACAUGGCGAAACUGAGCUGCAGAGCAUUGCGCAGUUCGUUGAGCACCAUGG ((.--------((-------......))..))....(((..-...----)))-...(((((.(.(((.(((((((..........))))))))))...).))))). ( -24.00) >DroYak_CAF1 170288 94 + 1 GCUGUGGAGUCGC-------UGUCUCGCUCACUUUUGUGUCCCAU----CGC-UUACAUGGCAAAACUGAGCUGCAGAGCAUUGCGCAACUCGUUGAGCACCAUGG .((((((.((..(-------(((...(((((.(((((....((((----...-....))))))))).))))).)))).))..(((.(((....))).))))))))) ( -26.00) >DroAna_CAF1 200575 90 + 1 UCU--------GC-------CAUCUCGCUUGCUUUCGCAUC-GCUGGCUCGCUUCACAUGGCGAAGCUGAGCUGCAGGGCAUUCCUCAGCUCGUUCAGCACCAUGG ...--------.(-------(((......(((....)))..-(((((((((((......)))).))).((((((.(((.....)))))))))...))))...)))) ( -32.00) >consensus GCU________GC_______UGUCUCGCUCGCUUUCGUGUC_CAU____CGC_UCACAUGGCGAAACUGAGCUGCAGAGCAUUGCGCAGCUCGUUGAGCACCAUGG ........................................................(((((.(.(((.(((((((..........))))))))))...).))))). (-18.68 = -18.90 + 0.22)

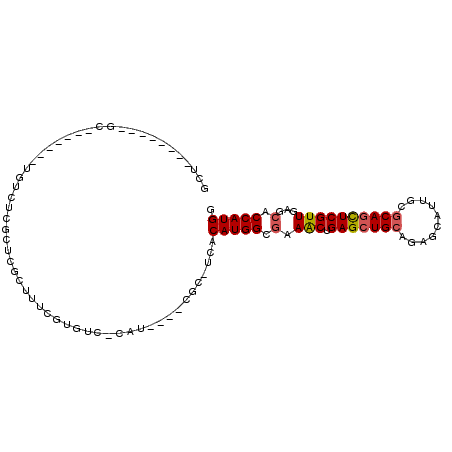
| Location | 4,381,841 – 4,381,933 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 78.34 |
| Mean single sequence MFE | -30.97 |
| Consensus MFE | -20.09 |
| Energy contribution | -20.32 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.871440 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4381841 92 - 22224390 CCAUGGUGCUCAACGAGCUGCGCAAUGCUCUGCAGCUCAGUUUCGCCAUGUGA-GCG----AUG-GACACGAAAGCGAACAAGACGAAAGCGAGC--------AGC ....(.(((((...((((((((........))))))))..(((((.(.(((..-((.----.((-....))...))..))).).)))))..))))--------).) ( -31.50) >DroSec_CAF1 167007 76 - 1 CCAUGGUGCUCAACGAGCUGCGCAAUGCUCUGCAGCUCAGUUUCGCCAUGUGA-GCG--------------AAACCGAGCGAGACA-------GC--------AGG ((.((.(((((...((((........))))....((((.((((((((....).-)))--------------)))).))))))).))-------.)--------))) ( -31.40) >DroSim_CAF1 154502 76 - 1 CCAUGGUGCUCAACGAGCUGCGCAAUGCUCUGCAGCUCAGUUUCGCCAUGUGA-GCG--------------AAACCGAGCGAGACA-------GC--------AGG ((.((.(((((...((((........))))....((((.((((((((....).-)))--------------)))).))))))).))-------.)--------))) ( -31.40) >DroEre_CAF1 177850 85 - 1 CCAUGGUGCUCAACGAACUGCGCAAUGCUCUGCAGCUCAGUUUCGCCAUGUGA-GCG----AUG-GACACGAAUGCAAGCGAGACA-------GC--------AGC ....(.((((....((.(((((........))))).)).(((((((..((((.-.((----...-....))..)))).))))))))-------))--------).) ( -23.30) >DroYak_CAF1 170288 94 - 1 CCAUGGUGCUCAACGAGUUGCGCAAUGCUCUGCAGCUCAGUUUUGCCAUGUAA-GCG----AUGGGACACAAAAGUGAGCGAGACA-------GCGACUCCACAGC .......(((....((((((((((......))).(((((.((((((((((...-.).----))))....))))).)))))......-------)))))))...))) ( -29.70) >DroAna_CAF1 200575 90 - 1 CCAUGGUGCUGAACGAGCUGAGGAAUGCCCUGCAGCUCAGCUUCGCCAUGUGAAGCGAGCCAGC-GAUGCGAAAGCAAGCGAGAUG-------GC--------AGA .(((((((..(...(((((((((.....))).))))))..)..)))))))........((((.(-(.(((....)))..))...))-------))--------... ( -38.50) >consensus CCAUGGUGCUCAACGAGCUGCGCAAUGCUCUGCAGCUCAGUUUCGCCAUGUGA_GCG____AUG_GACACGAAAGCGAGCGAGACA_______GC________AGC .(((((((...(((((((((((........)))))))).))).)))))))........................................................ (-20.09 = -20.32 + 0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:09:53 2006