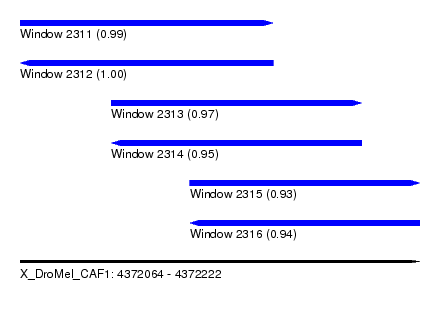
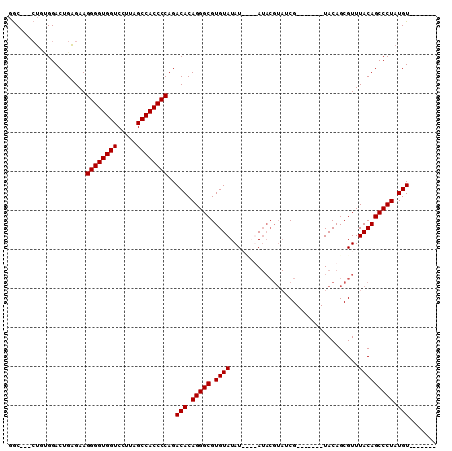
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,372,064 – 4,372,222 |
| Length | 158 |
| Max. P | 0.999228 |

| Location | 4,372,064 – 4,372,164 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 80.66 |
| Mean single sequence MFE | -35.68 |
| Consensus MFE | -28.49 |
| Energy contribution | -28.48 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.80 |
| SVM decision value | 2.29 |
| SVM RNA-class probability | 0.991894 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4372064 100 + 22224390 GGC---CUAAGGACUGAGAAGGGGUGGUCCUUAGCCACCCCAGACACAGGGCGUGUAUAUAUAUAUACGUAACGUACAGCGUACAGCGUUUACAGCCCUAUGU------- ...---..............((((((((.....))))))))..(((.(((((((((((....)))))).....(((.((((.....))))))).))))).)))------- ( -37.00) >DroSec_CAF1 157416 89 + 1 GGC---CUGUGGACUGAGAAGGGGUGGUCCUUAGCCACCCCAGACACAGGGCGUGUAUAU----AUACGUAUCG-------UACAGCGUUUACAGCCCUAUGU------- ((.---((((((((......((((((((.....))))))))....((...((((((....----))))))...)-------).....)))))))).)).....------- ( -34.40) >DroSim_CAF1 144928 89 + 1 GGC---CUGUGGACUGAGAAGGGGUGGUCCUUAGCCACCCCAGACACAGGGCGUGUAUAU----AUACGUAUCG-------UACAGCGUUUACAGCCCUAUGU------- ((.---((((((((......((((((((.....))))))))....((...((((((....----))))))...)-------).....)))))))).)).....------- ( -34.40) >DroEre_CAF1 168115 95 + 1 -ACGGGGGGCGGACUGCGAAGGGGUGGUCUUUAGCCACCCCAGACACAGGGCGUGUAUAUACAUA-------CG-------UACAGCGUUUACAGCCCUAUGUGAAGAGG -(((.(((((.((.(((...((((((((.....)))))))).......(..(((((......)))-------))-------..).))).))...))))).)))....... ( -36.90) >consensus GGC___CUGUGGACUGAGAAGGGGUGGUCCUUAGCCACCCCAGACACAGGGCGUGUAUAU____AUACGUAUCG_______UACAGCGUUUACAGCCCUAUGU_______ ....................((((((((.....))))))))..(((.(((((.((((.................................))))))))).)))....... (-28.49 = -28.48 + -0.00)


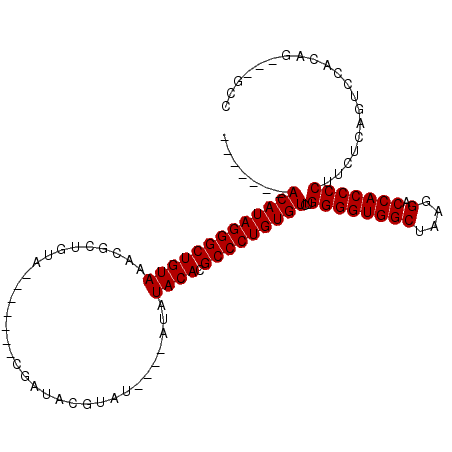
| Location | 4,372,064 – 4,372,164 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 80.66 |
| Mean single sequence MFE | -34.98 |
| Consensus MFE | -29.36 |
| Energy contribution | -29.36 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.19 |
| Structure conservation index | 0.84 |
| SVM decision value | 3.45 |
| SVM RNA-class probability | 0.999228 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4372064 100 - 22224390 -------ACAUAGGGCUGUAAACGCUGUACGCUGUACGUUACGUAUAUAUAUAUACACGCCCUGUGUCUGGGGUGGCUAAGGACCACCCCUUCUCAGUCCUUAG---GCC -------(((((((((((((...((.....))((((((...))))))......)))).)))))))))..((((((((....).)))))))......(((....)---)). ( -36.70) >DroSec_CAF1 157416 89 - 1 -------ACAUAGGGCUGUAAACGCUGUA-------CGAUACGUAU----AUAUACACGCCCUGUGUCUGGGGUGGCUAAGGACCACCCCUUCUCAGUCCACAG---GCC -------(((((((((((((.....((((-------((...)))))----)..)))).)))))))))..((((((((....).)))))))......(((....)---)). ( -34.70) >DroSim_CAF1 144928 89 - 1 -------ACAUAGGGCUGUAAACGCUGUA-------CGAUACGUAU----AUAUACACGCCCUGUGUCUGGGGUGGCUAAGGACCACCCCUUCUCAGUCCACAG---GCC -------(((((((((((((.....((((-------((...)))))----)..)))).)))))))))..((((((((....).)))))))......(((....)---)). ( -34.70) >DroEre_CAF1 168115 95 - 1 CCUCUUCACAUAGGGCUGUAAACGCUGUA-------CG-------UAUGUAUAUACACGCCCUGUGUCUGGGGUGGCUAAAGACCACCCCUUCGCAGUCCGCCCCCCGU- .......(((((((((((((.....((((-------(.-------...))))))))).)))))))))..((((((((....).)))))))...((.....)).......- ( -33.80) >consensus _______ACAUAGGGCUGUAAACGCUGUA_______CGAUACGUAU____AUAUACACGCCCUGUGUCUGGGGUGGCUAAGGACCACCCCUUCUCAGUCCACAG___GCC .......(((((((((((((.................................)))).)))))))))..((((((((....).))))))).................... (-29.36 = -29.36 + -0.00)


| Location | 4,372,100 – 4,372,199 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 81.79 |
| Mean single sequence MFE | -28.18 |
| Consensus MFE | -20.49 |
| Energy contribution | -20.11 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.966114 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
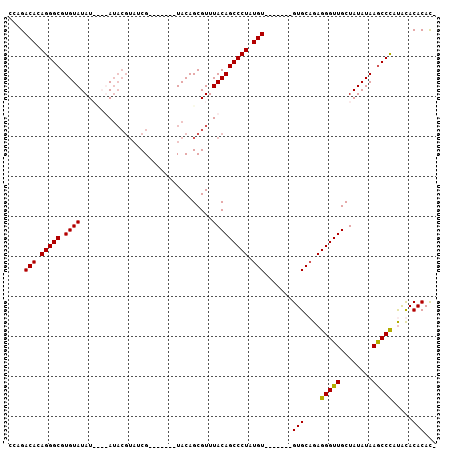
>X_DroMel_CAF1 4372100 99 + 22224390 CCAGACACAGGGCGUGUAUAUAUAUAUACGUAACGUACAGCGUACAGCGUUUACAGCCCUAUGU-------GUGCCGAGGGUUGCUAUAUAAGCCCAUGCACACAC- ........(((((((((((....)))))).....(((.((((.....))))))).))))).(((-------((((...(((((........)))))..))))))).- ( -36.10) >DroSec_CAF1 157452 88 + 1 CCAGACACAGGGCGUGUAUAU----AUACGUAUCG-------UACAGCGUUUACAGCCCUAUGU-------GUGCAGAGGGUUGCUAUAUAAGCCCAUAUACACAU- .((.(((.(((((.((((...----..((((....-------....)))).))))))))).)))-------.))....(((((........)))))..........- ( -27.20) >DroSim_CAF1 144964 88 + 1 CCAGACACAGGGCGUGUAUAU----AUACGUAUCG-------UACAGCGUUUACAGCCCUAUGU-------GUGCAGAGGGUUGCUAUAUAAGCCCAUAUACACAU- .((.(((.(((((.((((...----..((((....-------....)))).))))))))).)))-------.))....(((((........)))))..........- ( -27.20) >DroEre_CAF1 168153 93 + 1 CCAGACACAGGGCGUGUAUAUACAUA-------CG-------UACAGCGUUUACAGCCCUAUGUGAAGAGGGUGCAGAGGGUUGCUAUAUAAACCUACACACACACA ....(((.(((((.((((.......(-------((-------.....))).))))))))).))).......(((..(.(((((........))))).).)))..... ( -22.20) >consensus CCAGACACAGGGCGUGUAUAU____AUACGUAUCG_______UACAGCGUUUACAGCCCUAUGU_______GUGCAGAGGGUUGCUAUAUAAGCCCAUACACACAC_ ....(((.(((((.((((.................................))))))))).))).......(((....(((((........))))).....)))... (-20.49 = -20.11 + -0.38)

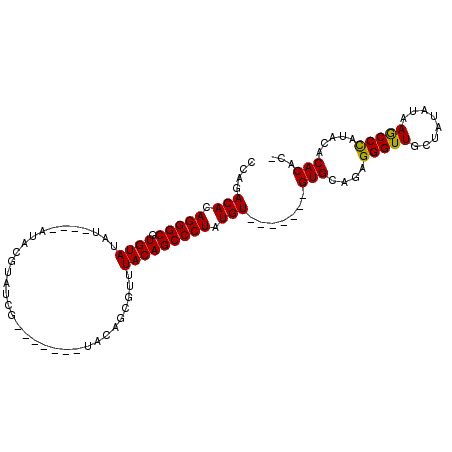
| Location | 4,372,100 – 4,372,199 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 81.79 |
| Mean single sequence MFE | -29.18 |
| Consensus MFE | -19.74 |
| Energy contribution | -20.11 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.945941 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
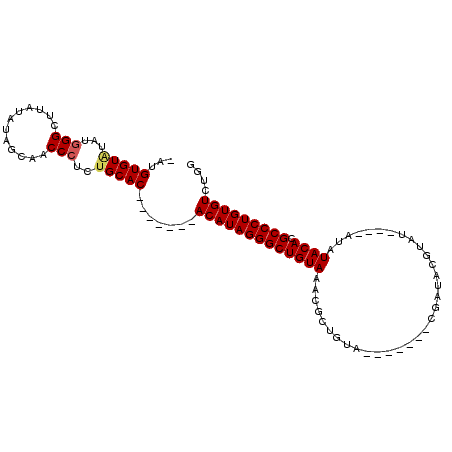
>X_DroMel_CAF1 4372100 99 - 22224390 -GUGUGUGCAUGGGCUUAUAUAGCAACCCUCGGCAC-------ACAUAGGGCUGUAAACGCUGUACGCUGUACGUUACGUAUAUAUAUAUACACGCCCUGUGUCUGG -..((((((..(((............)))...))))-------))(((((((((((...((.....))((((((...))))))......)))).)))))))...... ( -32.60) >DroSec_CAF1 157452 88 - 1 -AUGUGUAUAUGGGCUUAUAUAGCAACCCUCUGCAC-------ACAUAGGGCUGUAAACGCUGUA-------CGAUACGUAU----AUAUACACGCCCUGUGUCUGG -..(((((...(((............)))..)))))-------(((((((((((((.....((((-------((...)))))----)..)))).))))))))).... ( -28.30) >DroSim_CAF1 144964 88 - 1 -AUGUGUAUAUGGGCUUAUAUAGCAACCCUCUGCAC-------ACAUAGGGCUGUAAACGCUGUA-------CGAUACGUAU----AUAUACACGCCCUGUGUCUGG -..(((((...(((............)))..)))))-------(((((((((((((.....((((-------((...)))))----)..)))).))))))))).... ( -28.30) >DroEre_CAF1 168153 93 - 1 UGUGUGUGUGUAGGUUUAUAUAGCAACCCUCUGCACCCUCUUCACAUAGGGCUGUAAACGCUGUA-------CG-------UAUGUAUAUACACGCCCUGUGUCUGG .(((((((((((.((...((((((.......((((((((........)))).))))...))))))-------..-------.)).)))))))))))........... ( -27.50) >consensus _AUGUGUAUAUGGGCUUAUAUAGCAACCCUCUGCAC_______ACAUAGGGCUGUAAACGCUGUA_______CGAUACGUAU____AUAUACACGCCCUGUGUCUGG ...(((((...(((............)))..))))).......(((((((((((((.................................)))).))))))))).... (-19.74 = -20.11 + 0.37)


| Location | 4,372,131 – 4,372,222 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 87.22 |
| Mean single sequence MFE | -24.81 |
| Consensus MFE | -21.27 |
| Energy contribution | -20.50 |
| Covariance contribution | -0.77 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.925448 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
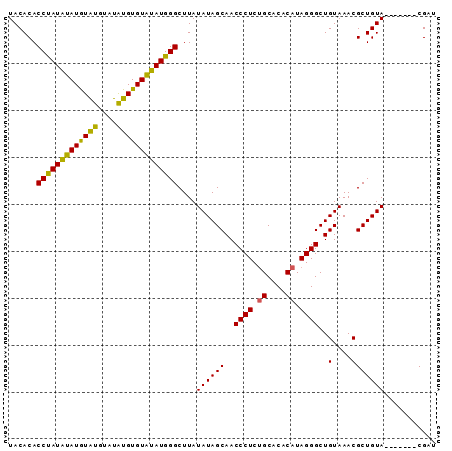
>X_DroMel_CAF1 4372131 91 + 22224390 AACGUACAGCGUACAGCGUUUACAGCCCUAUGUGUGCCGAGGGUUGCUAUAUAAGCCCAUGCACACACACACAUAUAUACAUAGGUGUGUA ...(((.((((.....)))))))........((((((...(((((........)))))..))))))((((((.(((....))).)))))). ( -28.90) >DroSec_CAF1 157479 84 + 1 AUCG-------UACAGCGUUUACAGCCCUAUGUGUGCAGAGGGUUGCUAUAUAAGCCCAUAUACACAUAUACAUACAUAUAUGGGUGUGUA ....-------...........(((((((.((....)).)))))))...((((.(((((((((..............))))))))).)))) ( -22.84) >DroSim_CAF1 144991 84 + 1 AUCG-------UACAGCGUUUACAGCCCUAUGUGUGCAGAGGGUUGCUAUAUAAGCCCAUAUACACAUAUACACACAUAUAUAGGUGUGUA ....-------....((.......))..((((((((....(((((........)))))...))))))))(((((((........))))))) ( -22.70) >consensus AUCG_______UACAGCGUUUACAGCCCUAUGUGUGCAGAGGGUUGCUAUAUAAGCCCAUAUACACAUAUACAUACAUAUAUAGGUGUGUA ....................((((..((((((((((....(((((........)))))(((......))).....))))))))))..)))) (-21.27 = -20.50 + -0.77)



| Location | 4,372,131 – 4,372,222 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 87.22 |
| Mean single sequence MFE | -24.37 |
| Consensus MFE | -21.84 |
| Energy contribution | -20.63 |
| Covariance contribution | -1.21 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938512 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4372131 91 - 22224390 UACACACCUAUGUAUAUAUGUGUGUGUGUGCAUGGGCUUAUAUAGCAACCCUCGGCACACAUAGGGCUGUAAACGCUGUACGCUGUACGUU (((((.(((((((((((((....)))))))))))))....((((((......((((.(......))))).....)))))).).)))).... ( -26.20) >DroSec_CAF1 157479 84 - 1 UACACACCCAUAUAUGUAUGUAUAUGUGUAUAUGGGCUUAUAUAGCAACCCUCUGCACACAUAGGGCUGUAAACGCUGUA-------CGAU ......((((((((..(((....)))..)))))))).(..((((((..((((.((....)).))))..(....)))))))-------..). ( -23.80) >DroSim_CAF1 144991 84 - 1 UACACACCUAUAUAUGUGUGUAUAUGUGUAUAUGGGCUUAUAUAGCAACCCUCUGCACACAUAGGGCUGUAAACGCUGUA-------CGAU .(((((........)))))(((((.((((.((((((((.....)))..((((.((....)).))))))))).))))))))-------)... ( -23.10) >consensus UACACACCUAUAUAUGUAUGUAUAUGUGUAUAUGGGCUUAUAUAGCAACCCUCUGCACACAUAGGGCUGUAAACGCUGUA_______CGAU ......(((((((((((((....)))))))))))))....((((((..((((.((....)).))))..(....)))))))........... (-21.84 = -20.63 + -1.21)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:09:45 2006