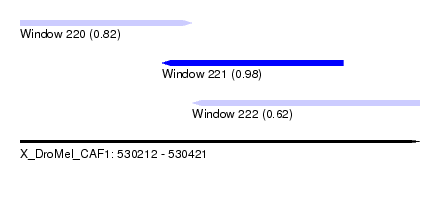
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 530,212 – 530,421 |
| Length | 209 |
| Max. P | 0.982438 |

| Location | 530,212 – 530,302 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.57 |
| Mean single sequence MFE | -22.64 |
| Consensus MFE | -16.22 |
| Energy contribution | -16.46 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.815656 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 530212 90 + 22224390 UGGACAACAUUGUGCCAGGCC------AACAAAAUCGUUAAUAACUGGCAUCUGGCACGUUCUUACCUGCAGCUAACAUGGCUUCCUUCACCA------------------------CCC .(((.......((((((((((------(.................))))..)))))))............((((.....))))))).......------------------------... ( -21.23) >DroSec_CAF1 16986 88 + 1 UGGACAACAUUGUGCCAGGCC------AACAAAAUAGUUAAUAACUGGCGUCUGGCAGGU--UUACCUGCACCUAACAUGGCAUCAUUUACCA------------------------CCC (((........((((((.(((------(.................)))).....(((((.--...)))))........))))))......)))------------------------... ( -21.97) >DroSim_CAF1 17265 88 + 1 UGGACAACAUUGUGCCAGGCC------AACAAAAUAGUUAAUAACUGGCGUCUGGCAGGU--UUACCUGCACCUAACAUGGCAUCAUUUACCA------------------------CCC (((........((((((.(((------(.................)))).....(((((.--...)))))........))))))......)))------------------------... ( -21.97) >DroEre_CAF1 21931 114 + 1 GGGACAACAUUGUGCCAGGCC------AACAAAAUCGUUAAUAACUGGCAUCUGGCACGUGCUUACCUGCAGCUAACAUGGCUUCCUCCACCACCACAACACCACAACACUCCAACACCC (((........((((((((((------(.................))))..)))))))((((......))((((.....))))............))....................))) ( -24.13) >DroYak_CAF1 16318 92 + 1 GGGACAACAUUGUGCCGGGCCAACGCCAACAAAAUCGUUAAUAACUGGCAUCUGGCACGUGCUUACCUGCAGCUAACAUGGCUUCCUCCACC---------------------------- ((((.......(((((((((((.....(((......)))......))))..))))))).(((......)))(((.....))).)))).....---------------------------- ( -23.90) >consensus UGGACAACAUUGUGCCAGGCC______AACAAAAUCGUUAAUAACUGGCAUCUGGCACGU_CUUACCUGCAGCUAACAUGGCUUCCUUCACCA________________________CCC .((.......(((((((((((......(((......))).......)))..))))))))......((((.......)).)).........))............................ (-16.22 = -16.46 + 0.24)

| Location | 530,286 – 530,381 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 86.14 |
| Mean single sequence MFE | -33.05 |
| Consensus MFE | -24.66 |
| Energy contribution | -25.29 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.21 |
| Mean z-score | -3.01 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.91 |
| SVM RNA-class probability | 0.982438 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 530286 95 - 22224390 ACAUGUUUCCUUGCCCCGCUUUUUUAUAGCCAUCUUCCUUCAAAAACAGAGGGUUAAAAGGGGUUGUAUGGAGGGGUUGGGGUGGUGAAGGAAGC ....(((((((((((((((((((((((((((.(((((((((.......)))))....)))))))))))..)))))).))))))....)))))))) ( -34.60) >DroSec_CAF1 17058 95 - 1 AAAUGCUUCCUUGGCCCGCCUUUUUAUGGCCAUCUUCCUUCGAAAAGAGAGGGUUAAAAGGGGUUGUAUGGAGGUGUUGGGGUGGUAAAUGAUGC ...((((.(((..((..(((((..((..(((.(((((((((.......)))))....)))))))..))..)))))))..))).))))........ ( -34.10) >DroSim_CAF1 17337 95 - 1 AAAUGUUUCCUUGGCCCGCCUUUUUAUGGCCAUCUUCCUUCGAAAAGAGAGGGUUAAAAGGGGUUGUAUGGAGGUGUUGGGGUGGUAAAUGAUGC ........(((..((..(((((..((..(((.(((((((((.......)))))....)))))))..))..)))))))..)))............. ( -29.70) >DroYak_CAF1 16398 89 - 1 AAAUGUU-CUUUGCCCCGGCUUUUUAUGGCCAUCUUCCUGCGAAAAGAGAGGGUUAAAAGGGGUUGUAUGUGGCUGUG-----GGUGGAGGAAGC .....((-(((..((((((((...((..(((((((((((......)).)))))).......)))..))...))))).)-----))..)))))... ( -33.81) >consensus AAAUGUUUCCUUGCCCCGCCUUUUUAUGGCCAUCUUCCUUCGAAAAGAGAGGGUUAAAAGGGGUUGUAUGGAGGUGUUGGGGUGGUAAAGGAAGC ....((((((((..(((((((((.(((((((.(((((((((.......)))))....))))))))))).))))))).......))..)))))))) (-24.66 = -25.29 + 0.63)


| Location | 530,302 – 530,421 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.46 |
| Mean single sequence MFE | -36.26 |
| Consensus MFE | -28.00 |
| Energy contribution | -27.84 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.619708 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 530302 119 - 22224390 AACUUUCUCCCAAAUGUGCCACUAAUUUGUGGGCAUGUGAACAUGUUU-CCUUGCCCCGCUUUUUUAUAGCCAUCUUCCUUCAAAAACAGAGGGUUAAAAGGGGUUGUAUGGAGGGGUUG .......((((.......((((......))))(((((....))))).(-((..((((((((.......)))......(((((.......)))))......))))).....)))))))... ( -35.60) >DroSec_CAF1 17074 119 - 1 AACUUACUCCCAAAUGUGCCACUAAUUUGUGGGCAUGUGAAAAUGCUU-CCUUGGCCCGCCUUUUUAUGGCCAUCUUCCUUCGAAAAGAGAGGGUUAAAAGGGGUUGUAUGGAGGUGUUG ..........((.((((.((((......)))))))).))..(((((((-((.(((((((((.......)))......(((((.......))))).......))))))...))))))))). ( -36.50) >DroSim_CAF1 17353 119 - 1 AACUUACUCCCAAAUGUGCCACUAAUUUGUGGGCAUGUGAAAAUGUUU-CCUUGGCCCGCCUUUUUAUGGCCAUCUUCCUUCGAAAAGAGAGGGUUAAAAGGGGUUGUAUGGAGGUGUUG .........(((((((((((((......)).)))))))(((.....))-).))))..((((((..((..(((.(((((((((.......)))))....)))))))..))..))))))... ( -34.30) >DroEre_CAF1 22045 120 - 1 AACUUUCUCCCAAAUGUGCCACUAAUUUGUGGGCAUGUGAAAAUGUUUCCUUUGGCCCGGCUUUUUAUGGCCAUUUUCCUUCGAAAAGAGAGGGUUAAAAGGGGUUGUAUGGGGGUGUUG (((...((((((.(((((((((......)).)))))))........((((((((((((((((......)))).(((((....)))))....))))).))))))).....)))))).))). ( -40.00) >DroYak_CAF1 16410 117 - 1 AACUUUCUCCCAAAUGUGCCACUAAUUUGUGGGCAUGUGAAAAUGUU--CUUUGCCCCGGCUUUUUAUGGCCAUCUUCCUGCGAAAAGAGAGGGUUAAAAGGGGUUGUAUGUGGCUGUG- .................(((((........((((((......)))))--)...(((((((((......))))((((((((......)).)))))).....))))).....)))))....- ( -34.90) >consensus AACUUUCUCCCAAAUGUGCCACUAAUUUGUGGGCAUGUGAAAAUGUUU_CCUUGGCCCGCCUUUUUAUGGCCAUCUUCCUUCGAAAAGAGAGGGUUAAAAGGGGUUGUAUGGAGGUGUUG ..........((.((((.((((......)))))))).))..................((((((..(((((((.(((((((((.......)))))....)))))))))))..))))))... (-28.00 = -27.84 + -0.16)

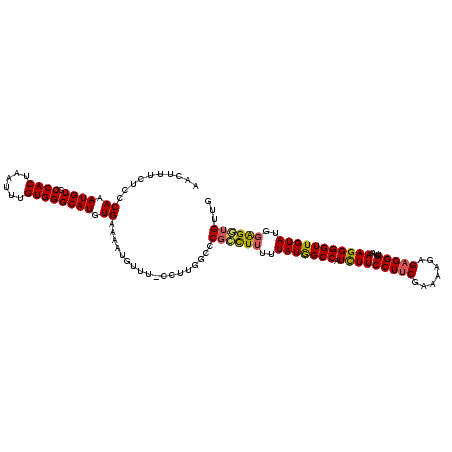
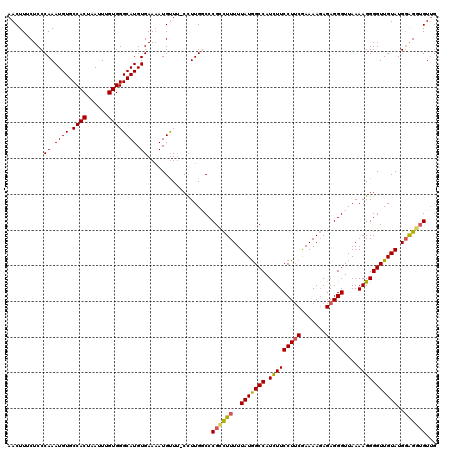
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:36:34 2006