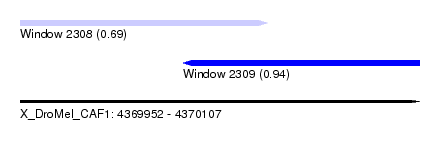
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,369,952 – 4,370,107 |
| Length | 155 |
| Max. P | 0.943641 |

| Location | 4,369,952 – 4,370,048 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 96.01 |
| Mean single sequence MFE | -19.59 |
| Consensus MFE | -18.40 |
| Energy contribution | -18.21 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.688657 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
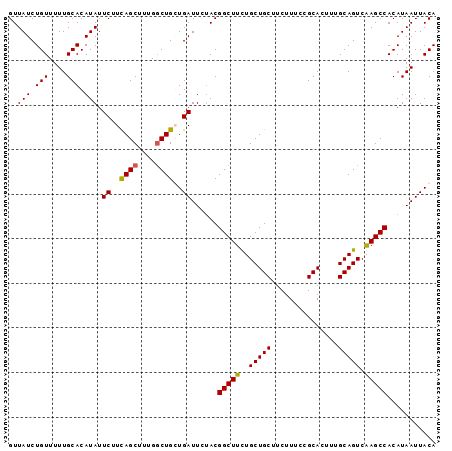
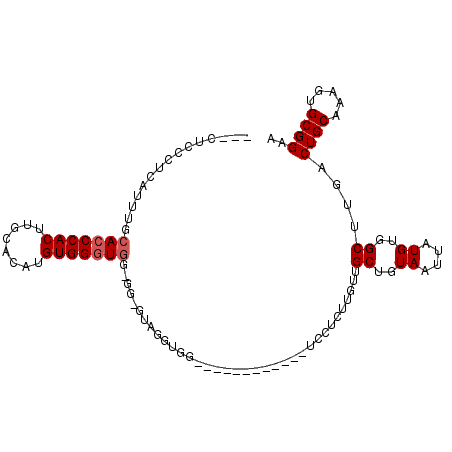
>X_DroMel_CAF1 4369952 96 + 22224390 GUUAUCUGUUUUUGCACAUAUUCUUUAGCUUUGGCUGCUGAUUCUACGGCUUUUGCUGCUUCUUUCCGCACUUUGCAGUCAAGCCACAUAAUUACA ..(((.(((....))).))).((..((((....))))..))......(((((..(((((...............))))).)))))........... ( -18.46) >DroSec_CAF1 155331 96 + 1 GUUAUCUGUUUUUGCACAUAUUCUUUAGCUUUGGCUGCUGAUUCUACGGCUUCUGCUGCUUCUUUCCGCACUUUGCAGUCAAGCCACAUAAUUACA ..(((.(((....))).))).((..((((....))))..))......((((((((((((........)))....))))..)))))........... ( -18.50) >DroEre_CAF1 165997 96 + 1 GUUAUCUGUUUUUGCACAUAUUCUUCAGCUUUGGCUGGUGAUUUUACGGCUUCUGCUGCUUCUUUCCGCACUUUGCAGUCGAGCCACAUAAUUACA ..(((.(((....))).))).....((((....))))((((((....((((((((((((........)))....))))..)))))....)))))). ( -23.50) >DroYak_CAF1 157626 96 + 1 GUUAUCUGUUUUUGCACAUAUUCUUCAGCUUUGUCUGGUGAUUCAACGGCUUCUGCUGCUUCUUUCCGCACUUUGCAGUCAAGCCACAUAAUUACA ..(((.(((....))).))).....(((......)))((((((....((((((((((((........)))....))))..)))))....)))))). ( -17.90) >consensus GUUAUCUGUUUUUGCACAUAUUCUUCAGCUUUGGCUGCUGAUUCUACGGCUUCUGCUGCUUCUUUCCGCACUUUGCAGUCAAGCCACAUAAUUACA ..(((.(((....))).))).((..((((....))))..))......(((((..(((((...............))))).)))))........... (-18.40 = -18.21 + -0.19)


| Location | 4,370,015 – 4,370,107 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 72.61 |
| Mean single sequence MFE | -29.76 |
| Consensus MFE | -16.44 |
| Energy contribution | -17.04 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.55 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.943641 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4370015 92 - 22224390 ---CUUCCUCAUUUGCACCCACAUGCACAUGUGGCUGGUGGAGUAGGUGG------------UCCUCUUGUUGCUGUAAUUAUGUGGCUUGACUGCAAAGUGCGGAA ---..(((.((((((((.(((((((.(((.(..((.((.(((.(....).------------))).)).))..))))...)))))))......))).))))).))). ( -28.60) >DroSec_CAF1 155394 104 - 1 ---CUCCCUCAUUUGCACCCACUUGCACAUGUGGGUGGUGGAGUAGGUGGUGGGGCUGGUGCUCCUCUUGUUGCUGUAAUUAUGUGGCUUGACUGCAAAGUGCGGAA ---...((.(((((.(((((((........))))))).....((((.....(((((....)))))((..((..(.........)..))..)))))).))))).)).. ( -36.20) >DroSim_CAF1 142884 104 - 1 ---CUCCCUCAUUUGCACCCACUUGCACAUGUGGGUUGAGGGGUAGGUGGUGGGGCUGGUGCUCCUCUUGUUGCUGUAAUUAUGUGGCUGNNNNNNNNNNNNNNNNN ---.(((((((.....((((((........))))))))))))).....((.(((((....))))).))....((..((....))..))................... ( -31.70) >DroEre_CAF1 166060 80 - 1 ---CUCUCCCAUUUUCACCCACUUACACAUGUGGGUGG------------------------UCCUCUUGUUGCUGUAAUUAUGUGGCUCGACUGCAAAGUGCGGAA ---...((((((((((((((((........)))))))(------------------------(..((..((..(.........)..))..))..)))))))).))). ( -25.70) >DroYak_CAF1 157689 83 - 1 CCUCUCUCCCAUUUUCACCCACUUACACAUGUGGGUGG------------------------UCCUCUUGUUGCUGUAAUUAUGUGGCUUGACUGCAAAGUGCGGAA ......((((((((((((((((........)))))))(------------------------(..((..((..(.........)..))..))..)))))))).))). ( -26.60) >consensus ___CUCCCUCAUUUGCACCCACUUGCACAUGUGGGUGG_GG_GUAGGUGG____________UCCUCUUGUUGCUGUAAUUAUGUGGCUUGACUGCAAAGUGCGGAA ...............(((((((........)))))))...................................((..((....))..))....((((.....)))).. (-16.44 = -17.04 + 0.60)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:09:40 2006