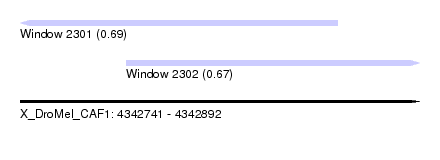
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,342,741 – 4,342,892 |
| Length | 151 |
| Max. P | 0.694273 |

| Location | 4,342,741 – 4,342,861 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.08 |
| Mean single sequence MFE | -28.88 |
| Consensus MFE | -24.74 |
| Energy contribution | -25.94 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.694273 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4342741 120 - 22224390 AAAACGUGUUAAAGGCGAUUUGGUGACAUCUCAUAGGAAAAUUAACUGGAGUCACCUCCGAAAUGUGCCAAGUCAACAUUUCACUCUGAACGGAGAUGUCAGAACUAUUAAUGAUGCAUA .....((((((...((......))((((((((...(((.........((((....))))(((((((.........)))))))..))).....))))))))...........))))))... ( -29.60) >DroSec_CAF1 128567 120 - 1 AAAACGUGUUAAAGGCGGUUUGGUGACAUCUCAUAGGAAAAUUAACUGGAGUCACCUCCGAAAUGUGCCAAGUCAACAUUUCACUCUGAACGGGAAUGUCAGAACUAUUAAUGAUGCAUA .....((((((..((..((((..(((((((((...(((.........((((....))))(((((((.........)))))))..)))....))).))))))))))..))..))))))... ( -28.70) >DroSim_CAF1 121973 120 - 1 AAAACGUGUUAAAGGCGGUUUGGUGACAUCUCAUAGGAAAAUUAACUGGAGUCACCUCCGAAAUGUGCCAAGUCAACAUUUCACUCUGAACGGGGAUGUCAGAACUAUUAAUGAUGCAUA .....((((((..((..((((..(((((((((...(((.........((((....))))(((((((.........)))))))..))).....)))))))))))))..))..))))))... ( -33.00) >DroEre_CAF1 136727 120 - 1 AAAACGUGUUAAAGGCGACUUGGUGACAUCUCAAAGGAAAAUUAACUGGAGUCACCUCCAAAAUGUGCCAAGUCAACAUUUCACUCUGAACGGGGAUGUCAGAACUAUUAAUGAUGCAUA .....((((((.....((((((((.((((((....)))........(((((....)))))...)))))))))))..........(((((.((....)))))))........))))))... ( -28.60) >DroYak_CAF1 130633 119 - 1 UAAACGUGUUAAAGGCGGUUUGGUAACAUCUCU-AGAAAAAUUAGCUGGAGUCACCUCCAAAAUGUGCCAAGUCAACAUUUUACUCUGAACGGGGAUGUCAGUACUAUUAAUGAUGCAUA .....((((((..((((.(((((......((((-((.........))))))......)))))...)))).((((.((((....((((....))))))))..).))).....))))))... ( -24.50) >consensus AAAACGUGUUAAAGGCGGUUUGGUGACAUCUCAUAGGAAAAUUAACUGGAGUCACCUCCGAAAUGUGCCAAGUCAACAUUUCACUCUGAACGGGGAUGUCAGAACUAUUAAUGAUGCAUA .....((((((.....(((((..(((((((((...(((.........((((....))))(((((((.........)))))))..))).....)))))))))))))).....))))))... (-24.74 = -25.94 + 1.20)



| Location | 4,342,781 – 4,342,892 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.38 |
| Mean single sequence MFE | -25.95 |
| Consensus MFE | -16.70 |
| Energy contribution | -19.50 |
| Covariance contribution | 2.80 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.667233 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4342781 111 + 22224390 AAUGUUGACUUGGCACAUUUCGGAGGUGACUCCAGUUAAUUUUCCUAUGAGAUGUCACCAAAUCGCCUUUAACACGUUUUCAUUUUUUUCCCCAUU---------UUGUGUUUAAGGAAC ......((.((((.(((((((((((....))))((.........))..)))))))..)))).)).((((.((((((....................---------.)))))).))))... ( -27.70) >DroSec_CAF1 128607 111 + 1 AAUGUUGACUUGGCACAUUUCGGAGGUGACUCCAGUUAAUUUUCCUAUGAGAUGUCACCAAACCGCCUUUAACACGUUUUCAUUUCUUUCCCACUU---------UGCUGUUAAAGUAAA .........((((.(((((((((((....))))((.........))..)))))))..)))).....((((((((.((...................---------.)))))))))).... ( -22.95) >DroSim_CAF1 122013 111 + 1 AAUGUUGACUUGGCACAUUUCGGAGGUGACUCCAGUUAAUUUUCCUAUGAGAUGUCACCAAACCGCCUUUAACACGUUUUCUUUUCAUUCCCACUU---------CGCUGUUAAAGUAAC .........((((.(((((((((((....))))((.........))..)))))))..)))).....((((((((((....................---------)).)))))))).... ( -23.65) >DroEre_CAF1 136767 120 + 1 AAUGUUGACUUGGCACAUUUUGGAGGUGACUCCAGUUAAUUUUCCUUUGAGAUGUCACCAAGUCGCCUUUAACACGUUUUUACUUGUUUCCCCUUUUUGUACACAUGCUGUUGAAGUAAC ......(((((((.((((((..((((((((....)))).....))))..))))))..)))))))..(((((((((((.(.(((...............))).).))).)))))))).... ( -32.46) >DroYak_CAF1 130673 113 + 1 AAUGUUGACUUGGCACAUUUUGGAGGUGACUCCAGCUAAUUUUUCU-AGAGAUGUUACCAAACCGCCUUUAACACGUUUA---AUGUUU---UUUUUUCUACACAUGCUGUUGAAGUUAC .........((((.(((((((((((....))))).(((.......)-))))))))..)))).....((((((((.((...---.(((..---........)))...)))))))))).... ( -23.00) >consensus AAUGUUGACUUGGCACAUUUCGGAGGUGACUCCAGUUAAUUUUCCUAUGAGAUGUCACCAAACCGCCUUUAACACGUUUUCAUUUCUUUCCCACUU_________UGCUGUUAAAGUAAC .........((((.(((((((((((....))))((.........))..)))))))..)))).....((((((((..................................)))))))).... (-16.70 = -19.50 + 2.80)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:09:33 2006