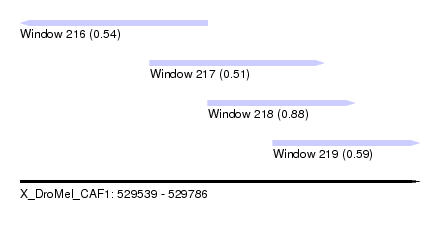
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 529,539 – 529,786 |
| Length | 247 |
| Max. P | 0.884728 |

| Location | 529,539 – 529,655 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.36 |
| Mean single sequence MFE | -23.66 |
| Consensus MFE | -20.52 |
| Energy contribution | -20.32 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.540909 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 529539 116 - 22224390 UAU----AAGUACCUUUAAUAACUUUACUUACACGAUGCUCGUUUAUUGAAAUUCCCACCCAUGGGAUACCACGCGCGUCGUCGUUCUUGACUUGACUUUUCCUCAACCCAUCUUGAAUG ..(----(((((.............)))))).(((((((.(((..........(((((....)))))....))).)))))))(((((..((.((((.......))))....))..))))) ( -22.26) >DroSec_CAF1 16259 116 - 1 UAU----AAGUACCUUUAAUAACUUUACUUACACGAUGCUCGUUUAUUGAAAUUUCCACCCAUGGGAUGCCUCGCGCGUCGUCGUUCUUGACUUGACUUUUCCUCAACUCAUCUUGAAUG ..(----(((((.............)))))).(((((((.((...........(((((....))))).....)).)))))))(((((.(((.((((.......)))).)))....))))) ( -20.51) >DroSim_CAF1 16573 116 - 1 UAU----AAGUACCUUUAAUAACUUUACUUACACGAUGCUCGUUUAUUGAAAUUUCCACCCAUGGGAUGCCUCGCGCGUCGUCGUUCUUGACUUGACUUUUCCUCAACUCAUCUUGAAUG ..(----(((((.............)))))).(((((((.((...........(((((....))))).....)).)))))))(((((.(((.((((.......)))).)))....))))) ( -20.51) >DroEre_CAF1 21249 120 - 1 UAUAUGUAAGUACCUUUAAUAACUUUACUUACACGAUGCUCGUUUAUUGAAAUUCCCACCCAUGGGAUGGCACACGCGUCGUCGUUCUUGACUUGACUUUGCCUCAACUCAUCUUGAAUG .....(((((((.............)))))))((((((((((.....)))...(((((....)))))........)))))))(((((.(((.((((.......)))).)))....))))) ( -26.82) >DroYak_CAF1 15680 120 - 1 UAUGUGUAAGUACCUUUAAUAACUUUACUUACGCGAUGCUCGUUUAUUGAAAUUCCCACCCAUGGGAUGGCACACGCGUCGUCGUUCUUGACUUGACUUUGCCUCAACUCAUCUUGAAUG ...(((((((((.............)))))))))((((((((.....)))...(((((....)))))........)))))..(((((.(((.((((.......)))).)))....))))) ( -28.22) >consensus UAU____AAGUACCUUUAAUAACUUUACUUACACGAUGCUCGUUUAUUGAAAUUCCCACCCAUGGGAUGCCACGCGCGUCGUCGUUCUUGACUUGACUUUUCCUCAACUCAUCUUGAAUG .......(((((.............)))))..((((((((((.....)))...(((((....)))))........)))))))(((((.(((.((((.......)))).)))....))))) (-20.52 = -20.32 + -0.20)

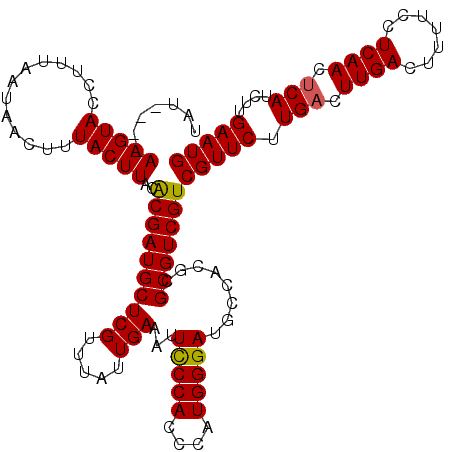
| Location | 529,619 – 529,727 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.58 |
| Mean single sequence MFE | -19.72 |
| Consensus MFE | -13.01 |
| Energy contribution | -14.81 |
| Covariance contribution | 1.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.66 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.505577 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 529619 108 + 22224390 AGCAUCGUGUAAGUAAAGUUAUUAAAGGUACUU----AUACAACCGAACUACCAGUAGCAACAGCAACAGCCCCACGGCAAUCC----AUAU----ACACAUAGUUACUUACUUACCCGU .....((.((((((((.((.((((..((((.((----.........)).)))).(((((....))....(((....))).....----...)----))...)))).)))))))))).)). ( -20.10) >DroSec_CAF1 16339 104 + 1 AGCAUCGUGUAAGUAAAGUUAUUAAAGGUACUU----AUACAACCGACCUACCAGCAGCAACAGCAACAGCCCCAGGGCAACCC----AUAU----ACACAUAGUUAC----UUACCCGU .....((.((((((((.(((......((((...----............))))....((....))))).(((....))).....----....----........))))----)))).)). ( -19.36) >DroSim_CAF1 16653 104 + 1 AGCAUCGUGUAAGUAAAGUUAUUAAAGGUACUU----AUACAACCGACCUACCAGUAGCAACAGCAACAGCCCCAGGGCAACCC----AUAU----ACACAUAGUUAC----UUACCCGU .....((.((((((((.((((((..((((....----.........))))...))))))..........(((....))).....----....----........))))----)))).)). ( -21.32) >DroEre_CAF1 21329 106 + 1 AGCAUCGUGUAAGUAAAGUUAUUAAAGGUACUUACAUAUACAACCCAACUACCAGUAGCU------ACAGCCCCACGGCAACCU----AUAU----ACACACAGUUACUUACUUACCCGU .....((.(((((((((((((((...((((...................)))))))))))------...(((....))).....----....----............)))))))).)). ( -18.31) >DroYak_CAF1 15760 114 + 1 AGCAUCGCGUAAGUAAAGUUAUUAAAGGUACUUACACAUACAACCCAACUACCAGUAGCA------ACAGCCCCACGGCAACCUCUGUAUAUACACACACACAGUUACUUACUUACCCGU .....((.((((((((..........((((...................)))).(((((.------...(((....)))......(((....)))........))))))))))))).)). ( -19.51) >consensus AGCAUCGUGUAAGUAAAGUUAUUAAAGGUACUU____AUACAACCGAACUACCAGUAGCAACAGCAACAGCCCCACGGCAACCC____AUAU____ACACAUAGUUACUUACUUACCCGU .....((.((((((((.((((((...((((...................))))))))))..........(((....))).............................)))))))).)). (-13.01 = -14.81 + 1.80)

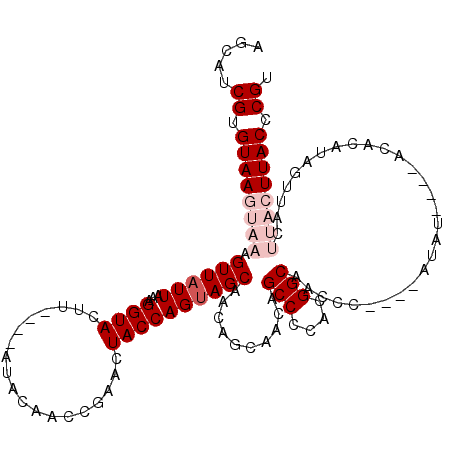
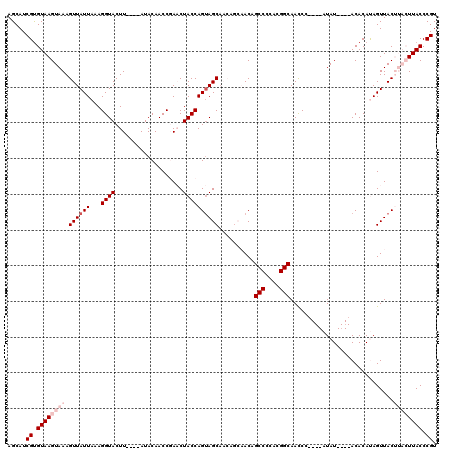
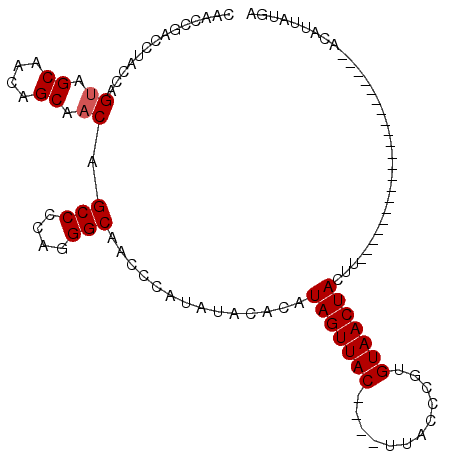
| Location | 529,655 – 529,746 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 80.71 |
| Mean single sequence MFE | -15.11 |
| Consensus MFE | -10.73 |
| Energy contribution | -11.07 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.884728 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 529655 91 + 22224390 CAACCGAACUACCAGUAGCAACAGCAACAGCCCCACGGCAAUCCAUAUACACAUAGUUACUUACUUACCCGUGUAACUACUU---------------------ACAUUAUGC ..............((.((....)).)).(((....)))....((((......(((((((............)))))))...---------------------....)))). ( -12.72) >DroSec_CAF1 16375 108 + 1 CAACCGACCUACCAGCAGCAACAGCAACAGCCCCAGGGCAACCCAUAUACACAUAGUUAC----UUACCCGUGUAACUACUUAUGAAUAUUUACAUUAUGAAUACAUUAUGA .................((....))....(((....)))....((((......(((((((----........)))))))..))))........(((.(((....))).))). ( -16.10) >DroSim_CAF1 16689 87 + 1 CAACCGACCUACCAGUAGCAACAGCAACAGCCCCAGGGCAACCCAUAUACACAUAGUUAC----UUACCCGUGUAACUACUU---------------------ACAUUAUGA ..............((.((....)).)).(((....)))....((((......(((((((----........)))))))...---------------------....)))). ( -16.52) >consensus CAACCGACCUACCAGUAGCAACAGCAACAGCCCCAGGGCAACCCAUAUACACAUAGUUAC____UUACCCGUGUAACUACUU_____________________ACAUUAUGA ..............((.((....)).)).(((....)))..............(((((((............)))))))................................. (-10.73 = -11.07 + 0.33)

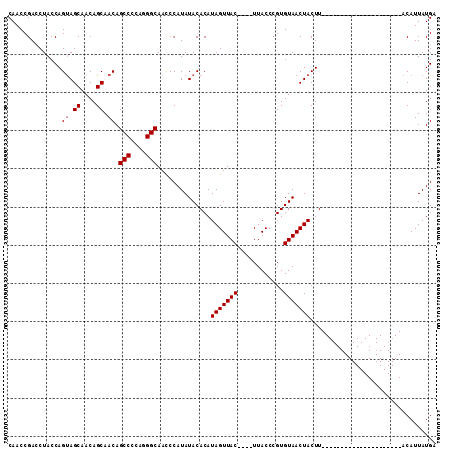
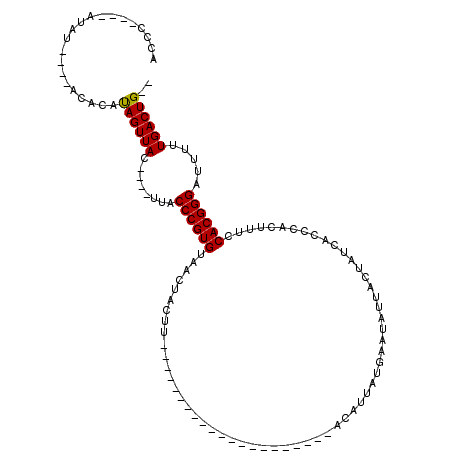
| Location | 529,695 – 529,786 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.42 |
| Mean single sequence MFE | -11.98 |
| Consensus MFE | -9.08 |
| Energy contribution | -8.89 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.594621 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 529695 91 + 22224390 AUCC----AUAU----ACACAUAGUUACUUACUUACCCGUGUAACUACUU---------------------ACAUUAUGCAUAUUACUAUCACCCACUUUCCACGGGAUUUUUGACUGUU ....----....----....(((((((........((((((.........---------------------..............................)))))).....))))))). ( -10.25) >DroSec_CAF1 16415 106 + 1 ACCC----AUAU----ACACAUAGUUAC----UUACCCGUGUAACUACUUAUGAAUAUUUACAUUAUGAAUACAUUAUGAAUAUUACUAUCACCCACUUUCCACGGGAUUUUUGACUG-- .(((----....----.....(((((((----........)))))))......((((((((....(((....)))..))))))))...................)))...........-- ( -15.50) >DroSim_CAF1 16729 85 + 1 ACCC----AUAU----ACACAUAGUUAC----UUACCCGUGUAACUACUU---------------------ACAUUAUGAAUAUUACUAUCACCCACUUUCCACGGGAUUUUUGACUG-- ...(----(((.----.....(((((((----........)))))))...---------------------....)))).............(((.........)))...........-- ( -11.62) >DroYak_CAF1 15834 97 + 1 ACCUCUGUAUAUACACACACACAGUUACUUACUUACCCGUGUAACUACUU---------------------ACAUUAUGCAUAUUACUAUCACCCACUUUCCACGGGAUUUUUGACUG-- .....................((((((........((((((.........---------------------..............................)))))).....))))))-- ( -10.55) >consensus ACCC____AUAU____ACACAUAGUUAC____UUACCCGUGUAACUACUU_____________________ACAUUAUGAAUAUUACUAUCACCCACUUUCCACGGGAUUUUUGACUG__ .....................((((((........((((((............................................................)))))).....)))))).. ( -9.08 = -8.89 + -0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:36:30 2006