| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,335,701 – 4,335,794 |
| Length | 93 |
| Max. P | 0.840000 |

| Location | 4,335,701 – 4,335,794 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 80.25 |
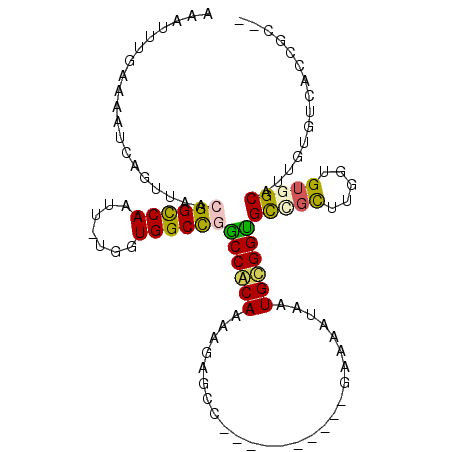
| Mean single sequence MFE | -29.65 |
| Consensus MFE | -19.30 |
| Energy contribution | -18.77 |
| Covariance contribution | -0.53 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.840000 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
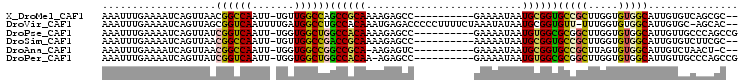
>X_DroMel_CAF1 4335701 93 + 22224390 AAAUUUGAAAAUCAGUUAACGGCCAAUU-UGUUGGCCAGCCGCAAAAGAGCC----------GAAAAUAAUGCGGUGCCGCUUGGUGUGGCAUUGUGUCAGCGC-- ....((((....((((....((((((..-..)))))).((((((.(((.(((----------(.........))))....)))..))))))))))..))))...-- ( -29.80) >DroVir_CAF1 206240 102 + 1 AAAUUUGAAAAUCAGUUAGCGGUCAAUUUUGAUGGCCUGCCACAAAUGAGACCCCCUUUUCUAAAUAUAAUGCGGUGUU-UUUGGUGUGGCAUUGUGC-AGCAC-- ..............(((.(((((((.......)))))(((((((...(((((((.(...............).)).)))-))...)))))))....))-)))..-- ( -25.66) >DroPse_CAF1 143624 95 + 1 AAAUUUGAAAAUCAGUUAUCGGUCAAUU-UGGUGGCUGGCCACAAAAGAGCC----------GAAAAUAAUGUGGCGCGGCUUGGUGUGGCAUUGUUGCCCAGCCG ............((((((((((.....)-)))))))))((((((........----------........)))))).(((((.((.(..((...))..)))))))) ( -31.19) >DroSim_CAF1 114961 93 + 1 AAAUUUGAAAAUCAGUUAACGGCCAAUU-UGUUGGCCGACCGCAAAAGAGCC----------AAAAAUAAUGCGGUGCCGCUUGGUGUGGCAUUGUGUCUUCGC-- ...((((............(((((((..-..)))))))...((......)))----------)))....((((((((((((.....))))))))))))......-- ( -29.80) >DroAna_CAF1 146673 91 + 1 AAAUUUGAAAAUCAGUUAACGGCCAAUU-UGGUGGCCGGCCGCA-AAGAGUC----------GAAAAUAAUGCGGUGCCGCUUAGUGUGGCAUUGUCUAACU-C-- ...(((((...((.((...((((((...-...))))))...)).-..)).))----------)))......((((((((((.....))))))))))......-.-- ( -29.40) >DroPer_CAF1 145649 94 + 1 AAAUUUGAAAAUCAGUUAUCGGUCAAUU-UGGUGGCUGGCCACAA-AGAGCC----------GAAAAUAAUGUGGCGCGGCUUGGUGUGGCAUUGUUGCCCAGCCG ...................((((.....-.((..((..((((((.-.(((((----------(..............))))))..))))))...))..))..)))) ( -32.04) >consensus AAAUUUGAAAAUCAGUUAACGGCCAAUU_UGGUGGCCGGCCACAAAAGAGCC__________GAAAAUAAUGCGGUGCCGCUUGGUGUGGCAUUGUGUCACCGC__ ...................((((((.......))))))((((((..........................))))))(((((.....)))))............... (-19.30 = -18.77 + -0.53)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:09:27 2006