| Sequence ID | X_DroMel_CAF1 |
|---|---|
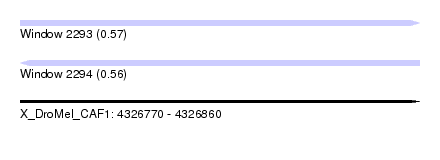
| Location | 4,326,770 – 4,326,860 |
| Length | 90 |
| Max. P | 0.569431 |

| Location | 4,326,770 – 4,326,860 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 76.76 |
| Mean single sequence MFE | -18.18 |
| Consensus MFE | -10.15 |
| Energy contribution | -12.02 |
| Covariance contribution | 1.86 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.569431 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
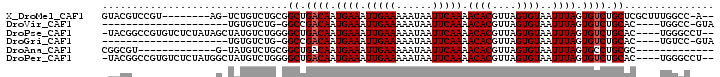
>X_DroMel_CAF1 4326770 90 + 22224390 --U-GGCCAAAGCGAGCAGACACUAAAUUACACUAACGUGUUUUGAAUUAUUUUUCAAUUUCAUUGUCAGCCGCAGACAGA-CU--------ACGGACGUAC --.-.......(((.((.((((.......((((....)))).(((((......)))))......)))).))))).......-(.--------...)...... ( -18.50) >DroVir_CAF1 167889 75 + 1 UAC-GGCCA----GUGCAGACACUAAAUUACACUAACGUGUUUUGAAUUAUUUUUCAAUUUCAUUGUCGGCC-CAGACACA--------------------- ...-(((((----(((....)))).....((((....)))).(((((......)))))..........))))-........--------------------- ( -16.90) >DroPse_CAF1 133751 95 + 1 --AGGCCCA----GUGCAGACACUAAAUUACACUAACGUGUUUUGAAUUAUUUUUCAAUUUCAUUGUCAGCCCCAGACAUAGCUAUAGAGACACGGCCGUA- --.((((.(----(((....)))).............((((((((((......)))........((((.......))))........)))))))))))...- ( -21.70) >DroGri_CAF1 133864 75 + 1 UAC-GGACA----GUGCAGACACUAAAUUACACUAACGUGUUUUGAAUUAUUUUUCAAUUUCAUUGUCGGCC-CAGACACA--------------------- ..(-.((((----(((.(((.........((((....)))).(((((......))))))))))))))).)..-........--------------------- ( -15.70) >DroAna_CAF1 134077 75 + 1 -------------GCGCAGGCACUAAAUUACACUAACGUGUUUUGAAUUAUUUUUCAAUUUCAUUGUCAGCCGCAGACAUA-C-------------ACGCCG -------------.....(((........((((....)))).(((((......)))))......((((.......))))..-.-------------..))). ( -14.60) >DroPer_CAF1 135535 95 + 1 --AGGCCCA----GUGCAGACACUAAAUUACACUAACGUGUUUUGAAUUAUUUUUCAAUUUCAUUGUCAGCCCCAGACAUAGCCAUAGAGACACGGCCGUA- --.((((.(----(((....)))).............((((((((((......)))........((((.......))))........)))))))))))...- ( -21.70) >consensus __A_GGCCA____GUGCAGACACUAAAUUACACUAACGUGUUUUGAAUUAUUUUUCAAUUUCAUUGUCAGCC_CAGACAUA_C______________CGUA_ ...............((.((((.......((((....)))).(((((......)))))......)))).))............................... (-10.15 = -12.02 + 1.86)

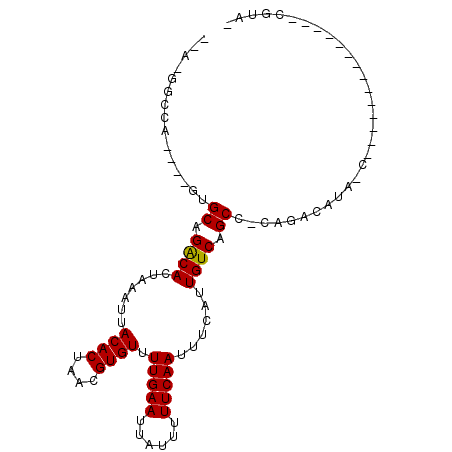

| Location | 4,326,770 – 4,326,860 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 76.76 |
| Mean single sequence MFE | -20.50 |
| Consensus MFE | -13.28 |
| Energy contribution | -13.45 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.561935 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4326770 90 - 22224390 GUACGUCCGU--------AG-UCUGUCUGCGGCUGACAAUGAAAUUGAAAAAUAAUUCAAAACACGUUAGUGUAAUUUAGUGUCUGCUCGCUUUGGCC-A-- .(((....))--------).-...(((.(((((.((((.((((.(((((......))))).((((....))))..)))).)))).)).)))...))).-.-- ( -21.80) >DroVir_CAF1 167889 75 - 1 ---------------------UGUGUCUG-GGCCGACAAUGAAAUUGAAAAAUAAUUCAAAACACGUUAGUGUAAUUUAGUGUCUGCAC----UGGCC-GUA ---------------------...(.(..-(((.((((.((((.(((((......))))).((((....))))..)))).)))).)).)----..).)-... ( -17.60) >DroPse_CAF1 133751 95 - 1 -UACGGCCGUGUCUCUAUAGCUAUGUCUGGGGCUGACAAUGAAAUUGAAAAAUAAUUCAAAACACGUUAGUGUAAUUUAGUGUCUGCAC----UGGGCCU-- -...((((((((......((((........))))((((.((((.(((((......))))).((((....))))..)))).)))).))))----..)))).-- ( -24.50) >DroGri_CAF1 133864 75 - 1 ---------------------UGUGUCUG-GGCCGACAAUGAAAUUGAAAAAUAAUUCAAAACACGUUAGUGUAAUUUAGUGUCUGCAC----UGUCC-GUA ---------------------...(..((-(((.((((.((((.(((((......))))).((((....))))..)))).)))).)).)----))..)-... ( -15.00) >DroAna_CAF1 134077 75 - 1 CGGCGU-------------G-UAUGUCUGCGGCUGACAAUGAAAUUGAAAAAUAAUUCAAAACACGUUAGUGUAAUUUAGUGCCUGCGC------------- .(((((-------------(-(.((((.......))))......(((((......))))).))))))).(((((..........)))))------------- ( -15.10) >DroPer_CAF1 135535 95 - 1 -UACGGCCGUGUCUCUAUGGCUAUGUCUGGGGCUGACAAUGAAAUUGAAAAAUAAUUCAAAACACGUUAGUGUAAUUUAGUGUCUGCAC----UGGGCCU-- -...(((((((....)))))))..(((..(.((.((((.((((.(((((......))))).((((....))))..)))).)))).)).)----..)))..-- ( -29.00) >consensus _UACG______________G_UAUGUCUG_GGCUGACAAUGAAAUUGAAAAAUAAUUCAAAACACGUUAGUGUAAUUUAGUGUCUGCAC____UGGCC_G__ ...............................((.((((.((((.(((((......))))).((((....))))..)))).)))).))............... (-13.28 = -13.45 + 0.17)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:09:26 2006