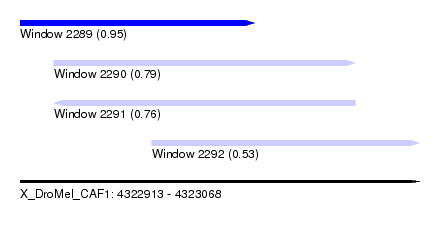
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,322,913 – 4,323,068 |
| Length | 155 |
| Max. P | 0.951642 |

| Location | 4,322,913 – 4,323,004 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 94.13 |
| Mean single sequence MFE | -32.50 |
| Consensus MFE | -27.93 |
| Energy contribution | -28.24 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.951642 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4322913 91 + 22224390 UCGAUGUUGCUGUUGCUGUUGCUGUUGCCAAUGCUGGCGUGUAAAGUGAACGGCGACGGCGGGAAAUCGUCAGCAAUGUCGCCGUAUUUUU .((((((((((((((((((((((..(((((....))))).....))).)))))))))(((((....))))))))))))))).......... ( -37.60) >DroSec_CAF1 109174 85 + 1 UCAAUGUUGCUGU------UGCUGUUGCCAAUGCUGGCGUGUAAAGUGAACGGCGACGGCGGGAAAUCGUCGGCAAUGUCGUCGUAUUUUU ......(((((((------(((((((((((....))))..........))))))))))))))(((((((.((((...)))).)).))))). ( -29.10) >DroSim_CAF1 102511 85 + 1 UCAAUGUUGCUGU------UGCUGUUGCCAAUGCUGGCGUGUAAAGUGAACGGCGACGGCGGGAAAUCGUCGGCAAUGUCGCCGUAUUUUU .....((((((((------((((..(((((....))))).....))).)))))))))(((((....)))))(((......)))........ ( -30.20) >DroEre_CAF1 116162 85 + 1 UCGAUGUUGCUGU------UGCUGUUGCCAUUGCUGGCGUGUAAAGUGAACGGCGACGGCGGGAAAUCGUCGGCAAUGUCGCCGUAUUUUU .((((((((((((------(((((((..(.((((......)))).)..)))))))))(((((....))))))))))))))).......... ( -33.10) >consensus UCAAUGUUGCUGU______UGCUGUUGCCAAUGCUGGCGUGUAAAGUGAACGGCGACGGCGGGAAAUCGUCGGCAAUGUCGCCGUAUUUUU .(((((((((((.......((((((((((..((((.........))))...)))))))))).........))))))))))).......... (-27.93 = -28.24 + 0.31)



| Location | 4,322,926 – 4,323,043 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 93.35 |
| Mean single sequence MFE | -39.28 |
| Consensus MFE | -32.96 |
| Energy contribution | -33.28 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.791038 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4322926 117 + 22224390 UGCUGUUGCUGUUGCCAAUGC--UGGCGUGUAAAGUGAACGGCGACGGCGGGAAAUCGUCAGCAAUGUCGCCGUAUUUUUUCGGGGUCCCUUACCUAGGCAAAUUUACGGUAAGGUAAU .((((((((((((((((....--))))..........))))))))))))((((...((.((....)).))(((........)))..))))((((((.(.(........).).)))))). ( -39.20) >DroSec_CAF1 109187 111 + 1 ------UGCUGUUGCCAAUGC--UGGCGUGUAAAGUGAACGGCGACGGCGGGAAAUCGUCGGCAAUGUCGUCGUAUUUUUUCGGGUUCCCUUACCUAGGCAAAUUUACGGUAAGGUAAU ------.(((((((((..(((--......)))..(....))))))))))(..(((.((.((((...)))).))...)))..)..(((.(((((((((((....)))).))))))).))) ( -36.50) >DroSim_CAF1 102524 111 + 1 ------UGCUGUUGCCAAUGC--UGGCGUGUAAAGUGAACGGCGACGGCGGGAAAUCGUCGGCAAUGUCGCCGUAUUUUUUCGGGGUCCCUUACCUAGGCAAAUUUACGGUAAGGUAAU ------.(((((((((..(((--......)))..(....))))))))))((((..(((.((((......))))........)))..))))((((((.(.(........).).)))))). ( -38.50) >DroEre_CAF1 116175 110 + 1 ------UGCUGUUGCCAUUGC--UGGCGUGUAAAGUGAACGGCGACGGCGGGAAAUCGUCGGCAAUGUCGCCGUAUUUUUUCGGGGUCCCUUACCUGGGCAAAUUUACGGUAAGGCA-A ------((((((((((.((((--......)))).(....)))))))(((((....))))))))).((((((((((..(((((.((((.....)))).)).)))..))))))..))))-. ( -39.30) >DroYak_CAF1 110783 113 + 1 ------UUCUGUUGCCAUUGCCGUGGCGUGUAAAGUGAACGGCGACGGCGGGAAAUCGUCGGCAAUGUCGCCGUAUUUUUUCGGGGUCCCUUACCUAGGCAAAUUCACGGUAAGGUAAA ------.....(((((.(((((((((..(((...((((((((((((.((.((......)).))...))))))))........((....))))))....)))...)))))))))))))). ( -42.90) >consensus ______UGCUGUUGCCAAUGC__UGGCGUGUAAAGUGAACGGCGACGGCGGGAAAUCGUCGGCAAUGUCGCCGUAUUUUUUCGGGGUCCCUUACCUAGGCAAAUUUACGGUAAGGUAAU .......(((((((((...((....))((.........)))))))))))(..(((....((((......))))...)))..)......(((((((((((....)))).))))))).... (-32.96 = -33.28 + 0.32)


| Location | 4,322,926 – 4,323,043 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 93.35 |
| Mean single sequence MFE | -29.83 |
| Consensus MFE | -28.44 |
| Energy contribution | -28.64 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.762584 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4322926 117 - 22224390 AUUACCUUACCGUAAAUUUGCCUAGGUAAGGGACCCCGAAAAAAUACGGCGACAUUGCUGACGAUUUCCCGCCGUCGCCGUUCACUUUACACGCCA--GCAUUGGCAACAGCAACAGCA ....(((((((.............)))))))..............((((((((...((.(........).)).))))))))...........(((.--.....)))....((....)). ( -30.92) >DroSec_CAF1 109187 111 - 1 AUUACCUUACCGUAAAUUUGCCUAGGUAAGGGAACCCGAAAAAAUACGACGACAUUGCCGACGAUUUCCCGCCGUCGCCGUUCACUUUACACGCCA--GCAUUGGCAACAGCA------ ....(((((((.............)))))))((((.((........))........(.(((((.........))))).))))).........(((.--.....))).......------ ( -24.22) >DroSim_CAF1 102524 111 - 1 AUUACCUUACCGUAAAUUUGCCUAGGUAAGGGACCCCGAAAAAAUACGGCGACAUUGCCGACGAUUUCCCGCCGUCGCCGUUCACUUUACACGCCA--GCAUUGGCAACAGCA------ ....(((((((.............)))))))..............((((((((...((.(........).)).))))))))...........(((.--.....))).......------ ( -29.72) >DroEre_CAF1 116175 110 - 1 U-UGCCUUACCGUAAAUUUGCCCAGGUAAGGGACCCCGAAAAAAUACGGCGACAUUGCCGACGAUUUCCCGCCGUCGCCGUUCACUUUACACGCCA--GCAAUGGCAACAGCA------ .-..(((((((.............)))))))..............((((((((...((.(........).)).))))))))...........((((--....)))).......------ ( -32.12) >DroYak_CAF1 110783 113 - 1 UUUACCUUACCGUGAAUUUGCCUAGGUAAGGGACCCCGAAAAAAUACGGCGACAUUGCCGACGAUUUCCCGCCGUCGCCGUUCACUUUACACGCCACGGCAAUGGCAACAGAA------ ....(((((((.............)))))))......((((.....(((((....)))))....))))..(((((.(((((..............))))).))))).......------ ( -32.16) >consensus AUUACCUUACCGUAAAUUUGCCUAGGUAAGGGACCCCGAAAAAAUACGGCGACAUUGCCGACGAUUUCCCGCCGUCGCCGUUCACUUUACACGCCA__GCAUUGGCAACAGCA______ ....(((((((.............)))))))..............((((((((...((.(........).)).))))))))...........((((......))))............. (-28.44 = -28.64 + 0.20)

| Location | 4,322,964 – 4,323,068 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 91.54 |
| Mean single sequence MFE | -32.20 |
| Consensus MFE | -26.22 |
| Energy contribution | -26.86 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.81 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.525056 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4322964 104 + 22224390 GGCGACGGCGGGAAAUCGUCAGCAAUGUCGCCGUAUUUUUUCGGGGUCCCUUACCUAGGCAAAUUUACGGUAAGGUAAUUUGCACAGAGAGAAAAACUAGAUA-A-- (((((((((((....)))))(....)))))))((.(((((((((....))....((..(((((((.((......)))))))))..)).)))))))))......-.-- ( -30.10) >DroSec_CAF1 109219 105 + 1 GGCGACGGCGGGAAAUCGUCGGCAAUGUCGUCGUAUUUUUUCGGGUUCCCUUACCUAGGCAAAUUUACGGUAAGGUAAUUUGCACAGAGAGAAAAACUAGGUAGG-- .((((((((((....)))))(((...))))))))........((....))(((((((((((((((.((......))))))))).(.....).....)))))))).-- ( -30.00) >DroSim_CAF1 102556 104 + 1 GGCGACGGCGGGAAAUCGUCGGCAAUGUCGCCGUAUUUUUUCGGGGUCCCUUACCUAGGCAAAUUUACGGUAAGGUAAUUUGCACAGAGAGAAAAACUAGGUA-A-- ((.(((..((..(((....((((......))))...)))..))..))))).((((((((((((((.((......))))))))).(.....).....)))))))-.-- ( -33.10) >DroEre_CAF1 116207 103 + 1 GGCGACGGCGGGAAAUCGUCGGCAAUGUCGCCGUAUUUUUUCGGGGUCCCUUACCUGGGCAAAUUUACGGUAAGGCA-AUUGCACUGAGCGAAAAUAUAGCCA-U-- (((..((((((....))))))((((((((((((((..(((((.((((.....)))).)).)))..))))))..))).-)))))................))).-.-- ( -36.20) >DroYak_CAF1 110817 106 + 1 GGCGACGGCGGGAAAUCGUCGGCAAUGUCGCCGUAUUUUUUCGGGGUCCCUUACCUAGGCAAAUUCACGGUAAGGUAAAUUGCACAGAGAGAAAAUAUAGCCA-CGC (((..((((((....))))))((((((.(.(((........))).).)(((((((.............)))))))...)))))................))).-... ( -31.62) >consensus GGCGACGGCGGGAAAUCGUCGGCAAUGUCGCCGUAUUUUUUCGGGGUCCCUUACCUAGGCAAAUUUACGGUAAGGUAAUUUGCACAGAGAGAAAAACUAGCUA_A__ ((((((.((.((......)).))...))))))...((((((((((........)))..(((((((..(.....)..)))))))...))))))).............. (-26.22 = -26.86 + 0.64)

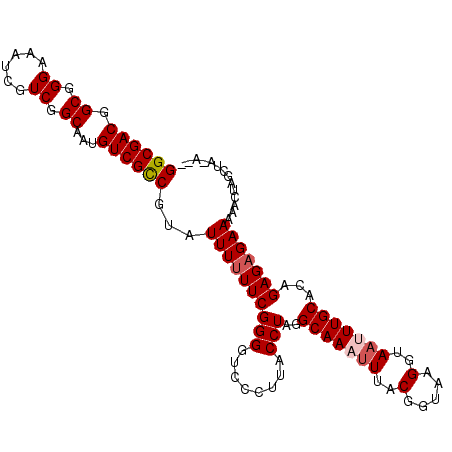

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:09:24 2006