
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,319,837 – 4,319,956 |
| Length | 119 |
| Max. P | 0.982068 |

| Location | 4,319,837 – 4,319,927 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 85.13 |
| Mean single sequence MFE | -24.23 |
| Consensus MFE | -20.18 |
| Energy contribution | -20.57 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.92 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.982068 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
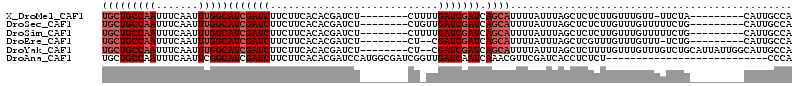
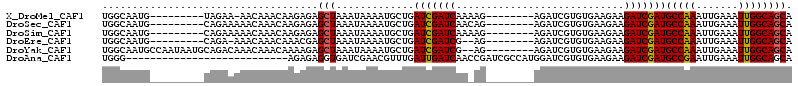
>X_DroMel_CAF1 4319837 90 + 22224390 -------------GAGAAACAUUUUGUUGAUCACUUGCUGCCAAUUUCAAUUUGGCAUCGAUCUUCUUCACACGAUCU--------CUUUUGAUCGAUCAGCAUUUUAUUU -------------...........((((((((..(((.((((((.......)))))).)))............((((.--------.....))))))))))))........ ( -24.20) >DroSec_CAF1 106202 90 + 1 -------------GAGAAACAUUUUGUUGAUCACUUGCUGCCAAUUUCAAUUUGGCAUCGAUCUUCUUCACACGAUCU--------CUGUUGAUCGAUCAGCAUUUUAUUU -------------...........((((((((..(((.((((((.......)))))).)))............((((.--------.....))))))))))))........ ( -24.20) >DroSim_CAF1 99571 90 + 1 -------------GAGAAACAUUUUGUUGAUCACUUGCUGCCAAUUUCAAUUUGGCAUCGAUCUUCUUCACACGAUCU--------CUUUUGAUCGAUCAGCAUUUUAUUU -------------...........((((((((..(((.((((((.......)))))).)))............((((.--------.....))))))))))))........ ( -24.20) >DroEre_CAF1 113222 88 + 1 -------------GCGAAACAUUUUGCUGAUCACUUGCUGCCAAUUUCAAUUUGGCAUCGAUCUUCUUCACACGAUCU--------CU--CGAUCGAUCAGCAUUUUAUUU -------------...........((((((((..(((.((((((.......)))))).)))............((((.--------..--.))))))))))))........ ( -25.80) >DroYak_CAF1 107932 88 + 1 -------------GAGAAACAUUUUGUUGAUCACUUGCUGCCAAUUUCAAUUUGGCAUCGAUCUUCUUCACACGAUCU--------CU--CGAUCGAUCAGCAUUUUAUUU -------------...........((((((((..(((.((((((.......)))))).)))............((((.--------..--.))))))))))))........ ( -23.40) >DroAna_CAF1 126130 111 + 1 AUUCAGCAAACGAGCUAAGCAUUUUGUUGAUCACUUGCUGCCAAUUUCAAUUCGGCAUCGAUCUUCUUCACACGAUCCAUGGCGAUCGGUUGAUCAAUCAAACGUUCGAUC ..........(((((.......(((((((((((.(((.(((((((....))).)))).)))...........(((((......)))))..))))))).)))).)))))... ( -23.60) >consensus _____________GAGAAACAUUUUGUUGAUCACUUGCUGCCAAUUUCAAUUUGGCAUCGAUCUUCUUCACACGAUCU________CU_UUGAUCGAUCAGCAUUUUAUUU ........................((((((((..(((.((((((.......)))))).)))............((((..............))))))))))))........ (-20.18 = -20.57 + 0.39)



| Location | 4,319,837 – 4,319,927 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 85.13 |
| Mean single sequence MFE | -21.65 |
| Consensus MFE | -16.31 |
| Energy contribution | -16.12 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.563930 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
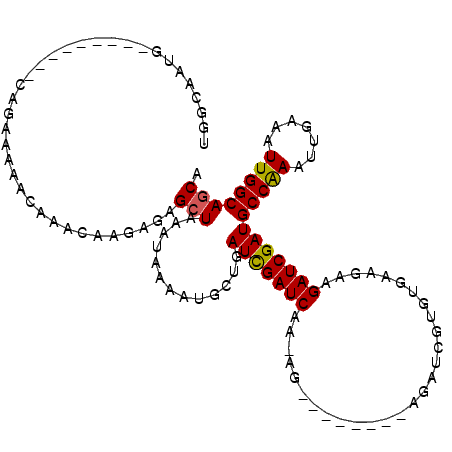
>X_DroMel_CAF1 4319837 90 - 22224390 AAAUAAAAUGCUGAUCGAUCAAAAG--------AGAUCGUGUGAAGAAGAUCGAUGCCAAAUUGAAAUUGGCAGCAAGUGAUCAACAAAAUGUUUCUC------------- ..........((...(((((.....--------.))))).....))..(((((.((((((.......)))))).....)))))...............------------- ( -17.80) >DroSec_CAF1 106202 90 - 1 AAAUAAAAUGCUGAUCGAUCAACAG--------AGAUCGUGUGAAGAAGAUCGAUGCCAAAUUGAAAUUGGCAGCAAGUGAUCAACAAAAUGUUUCUC------------- ..........((...(((((.....--------.))))).....))..(((((.((((((.......)))))).....)))))...............------------- ( -17.80) >DroSim_CAF1 99571 90 - 1 AAAUAAAAUGCUGAUCGAUCAAAAG--------AGAUCGUGUGAAGAAGAUCGAUGCCAAAUUGAAAUUGGCAGCAAGUGAUCAACAAAAUGUUUCUC------------- ..........((...(((((.....--------.))))).....))..(((((.((((((.......)))))).....)))))...............------------- ( -17.80) >DroEre_CAF1 113222 88 - 1 AAAUAAAAUGCUGAUCGAUCG--AG--------AGAUCGUGUGAAGAAGAUCGAUGCCAAAUUGAAAUUGGCAGCAAGUGAUCAGCAAAAUGUUUCGC------------- ........((((((((((((.--..--------.))))..............(.((((((.......)))))).)....))))))))...........------------- ( -26.90) >DroYak_CAF1 107932 88 - 1 AAAUAAAAUGCUGAUCGAUCG--AG--------AGAUCGUGUGAAGAAGAUCGAUGCCAAAUUGAAAUUGGCAGCAAGUGAUCAACAAAAUGUUUCUC------------- ..........((...(((((.--..--------.))))).....))..(((((.((((((.......)))))).....)))))...............------------- ( -20.30) >DroAna_CAF1 126130 111 - 1 GAUCGAACGUUUGAUUGAUCAACCGAUCGCCAUGGAUCGUGUGAAGAAGAUCGAUGCCGAAUUGAAAUUGGCAGCAAGUGAUCAACAAAAUGCUUAGCUCGUUUGCUGAAU (((((...((((((((..(((..(((((......)))))..)))....))))))((((((.......))))))))...)))))..........(((((......))))).. ( -29.30) >consensus AAAUAAAAUGCUGAUCGAUCAA_AG________AGAUCGUGUGAAGAAGAUCGAUGCCAAAUUGAAAUUGGCAGCAAGUGAUCAACAAAAUGUUUCUC_____________ (((((...((.((((((.................((((..........))))(.((((((.......)))))).)...)))))).))...)))))................ (-16.31 = -16.12 + -0.19)


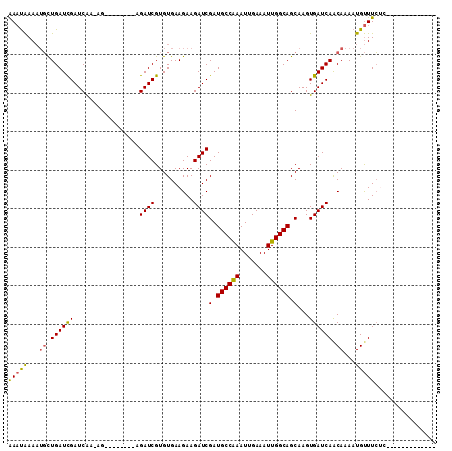
| Location | 4,319,859 – 4,319,956 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 80.72 |
| Mean single sequence MFE | -17.56 |
| Consensus MFE | -12.59 |
| Energy contribution | -13.42 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.815784 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4319859 97 + 22224390 UGCUGCCAAUUUCAAUUUGGCAUCGAUCUUCUUCACACGAUCU--------CUUUUGAUCGAUCAGCAUUUUAUUUAGCUCUCUUGUUUGUU-UUCUA---------CAUUGCCA (((((((((.......)))))................(((((.--------.....)))))...)))).................((.(((.-....)---------))..)).. ( -16.80) >DroSec_CAF1 106224 98 + 1 UGCUGCCAAUUUCAAUUUGGCAUCGAUCUUCUUCACACGAUCU--------CUGUUGAUCGAUCAGCAUUUUAUUUAGCUCUCUUGUUUGUUUUUCUG---------CAUUGCCA .(.((((((.......)))))).)((......))...(((((.--------.....)))))....(((........(((......)))........))---------)....... ( -17.59) >DroSim_CAF1 99593 98 + 1 UGCUGCCAAUUUCAAUUUGGCAUCGAUCUUCUUCACACGAUCU--------CUUUUGAUCGAUCAGCAUUUUAUUUAGCUCUCUUGUUUGUUUUUCUG---------CAUUGCCA .(.((((((.......)))))).)((......))...(((((.--------.....)))))....(((........(((......)))........))---------)....... ( -17.59) >DroEre_CAF1 113244 95 + 1 UGCUGCCAAUUUCAAUUUGGCAUCGAUCUUCUUCACACGAUCU--------CU--CGAUCGAUCAGCAUUUUAUUUAGCUCGUUUGUUUGUUU-UCUG---------CAUUGCCA (((((((((.......)))))(((((((....((....))...--------..--.))))))).)))).................((.(((..-...)---------))..)).. ( -17.10) >DroYak_CAF1 107954 105 + 1 UGCUGCCAAUUUCAAUUUGGCAUCGAUCUUCUUCACACGAUCU--------CU--CGAUCGAUCAGCAUUUUAUUUAGCUCUUUUGUUUGUUUGUCUGCAUUAUUGGCAUUGCCA (((((((((.......)))))(((((((....((....))...--------..--.))))))).))))....................(((......)))....((((...)))) ( -19.80) >DroAna_CAF1 126165 88 + 1 UGCUGCCAAUUUCAAUUCGGCAUCGAUCUUCUUCACACGAUCCAUGGCGAUCGGUUGAUCAAUCAAACGUUCGAUCACCUCUCU---------------------------CCCA .(.(((((((....))).)))).)((((..........))))...((.((((((((((....))))....)))))).)).....---------------------------.... ( -16.50) >consensus UGCUGCCAAUUUCAAUUUGGCAUCGAUCUUCUUCACACGAUCU________CU_UUGAUCGAUCAGCAUUUUAUUUAGCUCUCUUGUUUGUUUUUCUG_________CAUUGCCA (((((((((.......)))))(((((((............................))))))).))))............................................... (-12.59 = -13.42 + 0.83)
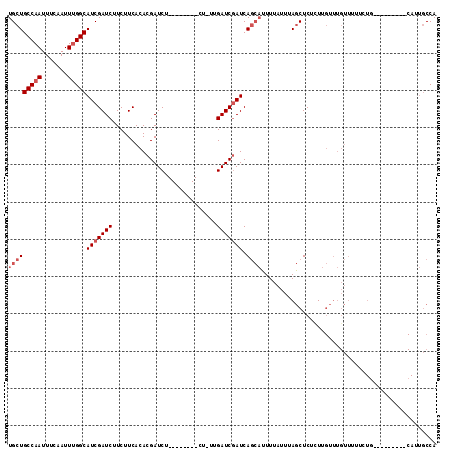
| Location | 4,319,859 – 4,319,956 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 80.72 |
| Mean single sequence MFE | -20.52 |
| Consensus MFE | -14.42 |
| Energy contribution | -14.31 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.837474 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
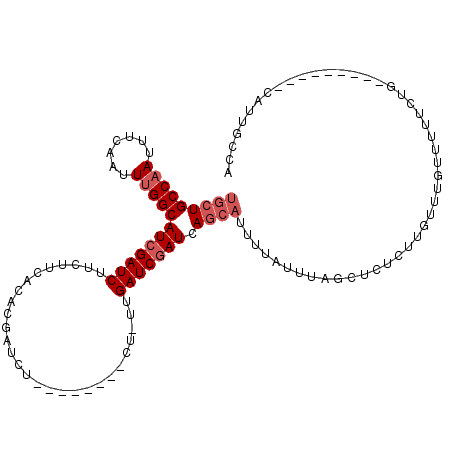
>X_DroMel_CAF1 4319859 97 - 22224390 UGGCAAUG---------UAGAA-AACAAACAAGAGAGCUAAAUAAAAUGCUGAUCGAUCAAAAG--------AGAUCGUGUGAAGAAGAUCGAUGCCAAAUUGAAAUUGGCAGCA ..((....---------.....-..((.((..((.(((..........)))..))((((.....--------.)))))).))...........((((((.......)))))))). ( -17.80) >DroSec_CAF1 106224 98 - 1 UGGCAAUG---------CAGAAAAACAAACAAGAGAGCUAAAUAAAAUGCUGAUCGAUCAACAG--------AGAUCGUGUGAAGAAGAUCGAUGCCAAAUUGAAAUUGGCAGCA ......((---------(.......((.((..((.(((..........)))..))((((.....--------.)))))).))...........((((((.......))))))))) ( -18.00) >DroSim_CAF1 99593 98 - 1 UGGCAAUG---------CAGAAAAACAAACAAGAGAGCUAAAUAAAAUGCUGAUCGAUCAAAAG--------AGAUCGUGUGAAGAAGAUCGAUGCCAAAUUGAAAUUGGCAGCA ......((---------(.......((.((..((.(((..........)))..))((((.....--------.)))))).))...........((((((.......))))))))) ( -18.00) >DroEre_CAF1 113244 95 - 1 UGGCAAUG---------CAGA-AAACAAACAAACGAGCUAAAUAAAAUGCUGAUCGAUCG--AG--------AGAUCGUGUGAAGAAGAUCGAUGCCAAAUUGAAAUUGGCAGCA ......((---------(...-...........(((((..........))....(((((.--..--------.)))))...........))).((((((.......))))))))) ( -19.70) >DroYak_CAF1 107954 105 - 1 UGGCAAUGCCAAUAAUGCAGACAAACAAACAAAAGAGCUAAAUAAAAUGCUGAUCGAUCG--AG--------AGAUCGUGUGAAGAAGAUCGAUGCCAAAUUGAAAUUGGCAGCA ..((..(((((((....((.((.............(((..........)))....((((.--..--------.)))))).)).......(((((.....))))).))))))))). ( -24.70) >DroAna_CAF1 126165 88 - 1 UGGG---------------------------AGAGAGGUGAUCGAACGUUUGAUUGAUCAACCGAUCGCCAUGGAUCGUGUGAAGAAGAUCGAUGCCGAAUUGAAAUUGGCAGCA ..((---------------------------.((..(((((((((........)))))).)))..)).))...((((..........))))(.((((((.......)))))).). ( -24.90) >consensus UGGCAAUG_________CAGAAAAACAAACAAGAGAGCUAAAUAAAAUGCUGAUCGAUCAA_AG________AGAUCGUGUGAAGAAGAUCGAUGCCAAAUUGAAAUUGGCAGCA ....................................(((.............(((((((............................)))))))(((((.......)))))))). (-14.42 = -14.31 + -0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:09:20 2006