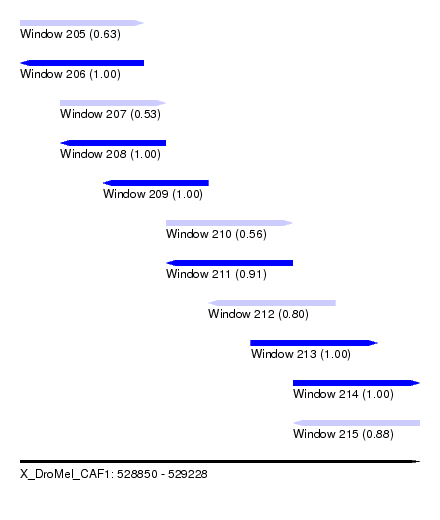
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 528,850 – 529,228 |
| Length | 378 |
| Max. P | 0.999904 |

| Location | 528,850 – 528,967 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.01 |
| Mean single sequence MFE | -30.56 |
| Consensus MFE | -26.84 |
| Energy contribution | -26.88 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.626375 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
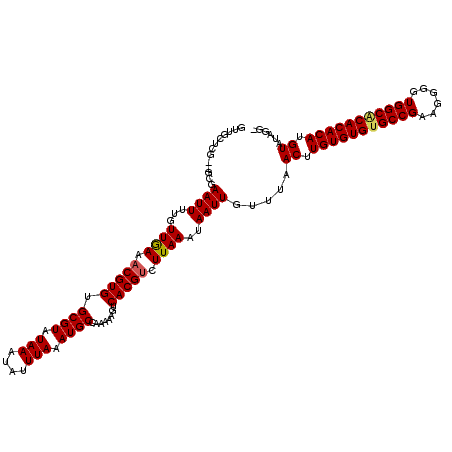
>X_DroMel_CAF1 528850 117 + 22224390 GUUGCUCG--GCGAAUUUUGUUGAAACGUGUGCGUAUAAAUAUUUAAAUGCCAAAAAGUCACGUCUUAAAUAAUUGUUUAACUUGUGUGCGCCGAAGGGGUGGCACACACAUGUAUAGG- .....(((--((((...))))))).(((((((.((.(((....)))...((((....((((((..((((((....))))))..)))).)).((....)).)))))).))))))).....- ( -28.00) >DroSec_CAF1 15568 117 + 1 GUUGCUCG--GCGAAUUUUGUUGAAACGUGUGCGUAUAAAUAUUUAAAUGCCAAAAAGUCACGGCUUAAAUAAUUGUUUAACUUGUGUGUGCCGAAGGGGUGGCACACACAUGUAUAGG- .....(((--((((...)))))))..(.(((((((((((.(((((((..(((..........)))))))))).)))).......((((((((((......))))))))))))))))).)- ( -33.60) >DroSim_CAF1 15887 117 + 1 GUUGCUCG--GCGAAUUUUGUUGAAACGUGUGCGUAUAAAUAUUUAAAUGCCAAAAAGUCACGUCUUAAAUAAUUGUUUAACUUGUGUGUGCCGAAGGGGUGGCACACACAUGUAUAGG- .....(((--((((...)))))))..(.(((((((((((.((((((((((...........))).))))))).)))).......((((((((((......))))))))))))))))).)- ( -29.60) >DroEre_CAF1 20571 119 + 1 GGUGCUUCAUCCGAAUUUUGUUAAAACGUGUGCGUAUAAAUAUUUAAAUGCCAAAAAGUCACGUCUUAAAUAAUUGUUUAACUUGUGUGUGCCGAAGGGGUGGCGCACACAUGUAUGGG- ..........(((((((...((((.(((((.((((.(((....))).))))........))))).))))..)))).....((.(((((((((((......))))))))))).)).))).- ( -29.90) >DroYak_CAF1 14996 117 + 1 G-UGCUCG--GCGAAUUUUGUUAAAACGUGUGCGUAUAAAUAUUUAAAUGCCAAAAAGUCACGUCUUAAAUAAUUGUUUAACUUGUGUGUGCCGAAGGGGUGGCACACACAUGUGUGGGU .-.(((((--((.((((...((((.(((((.((((.(((....))).))))........))))).))))..)))))))..((.(((((((((((......))))))))))).))..)))) ( -31.70) >consensus GUUGCUCG__GCGAAUUUUGUUGAAACGUGUGCGUAUAAAUAUUUAAAUGCCAAAAAGUCACGUCUUAAAUAAUUGUUUAACUUGUGUGUGCCGAAGGGGUGGCACACACAUGUAUAGG_ .............((((...((((.(((((.((((.(((....))).))))........))))).))))..)))).....((.(((((((((((......))))))))))).))...... (-26.84 = -26.88 + 0.04)

| Location | 528,850 – 528,967 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.01 |
| Mean single sequence MFE | -26.03 |
| Consensus MFE | -22.50 |
| Energy contribution | -22.78 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.86 |
| SVM decision value | 3.13 |
| SVM RNA-class probability | 0.998528 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 528850 117 - 22224390 -CCUAUACAUGUGUGUGCCACCCCUUCGGCGCACACAAGUUAAACAAUUAUUUAAGACGUGACUUUUUGGCAUUUAAAUAUUUAUACGCACACGUUUCAACAAAAUUCGC--CGAGCAAC -....(((.((((((((((........))))))))))..(((((......)))))...)))....((((((..............(((....))).............))--)))).... ( -24.43) >DroSec_CAF1 15568 117 - 1 -CCUAUACAUGUGUGUGCCACCCCUUCGGCACACACAAGUUAAACAAUUAUUUAAGCCGUGACUUUUUGGCAUUUAAAUAUUUAUACGCACACGUUUCAACAAAAUUCGC--CGAGCAAC -........((((((((((........)))))))))).(((((((...(((((((((((........))))..))))))).......(....))))).))).........--........ ( -26.60) >DroSim_CAF1 15887 117 - 1 -CCUAUACAUGUGUGUGCCACCCCUUCGGCACACACAAGUUAAACAAUUAUUUAAGACGUGACUUUUUGGCAUUUAAAUAUUUAUACGCACACGUUUCAACAAAAUUCGC--CGAGCAAC -....(((.((((((((((........))))))))))..(((((......)))))...)))....((((((..............(((....))).............))--)))).... ( -24.83) >DroEre_CAF1 20571 119 - 1 -CCCAUACAUGUGUGCGCCACCCCUUCGGCACACACAAGUUAAACAAUUAUUUAAGACGUGACUUUUUGGCAUUUAAAUAUUUAUACGCACACGUUUUAACAAAAUUCGGAUGAAGCACC -.((..((.((((((.(((........))).)))))).))...........((((((((((.(.....)((...(((....)))...)).))))))))))........)).......... ( -26.00) >DroYak_CAF1 14996 117 - 1 ACCCACACAUGUGUGUGCCACCCCUUCGGCACACACAAGUUAAACAAUUAUUUAAGACGUGACUUUUUGGCAUUUAAAUAUUUAUACGCACACGUUUUAACAAAAUUCGC--CGAGCA-C .........((((((((((........)))))))))).(((....((((..((((((((((.(.....)((...(((....)))...)).))))))))))...))))...--..))).-. ( -28.30) >consensus _CCUAUACAUGUGUGUGCCACCCCUUCGGCACACACAAGUUAAACAAUUAUUUAAGACGUGACUUUUUGGCAUUUAAAUAUUUAUACGCACACGUUUCAACAAAAUUCGC__CGAGCAAC ......((.((((((((((........)))))))))).))..............(((((((.(.....)((...(((....)))...)).)))))))....................... (-22.50 = -22.78 + 0.28)

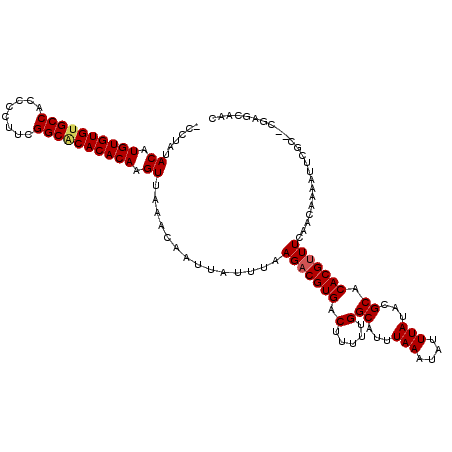
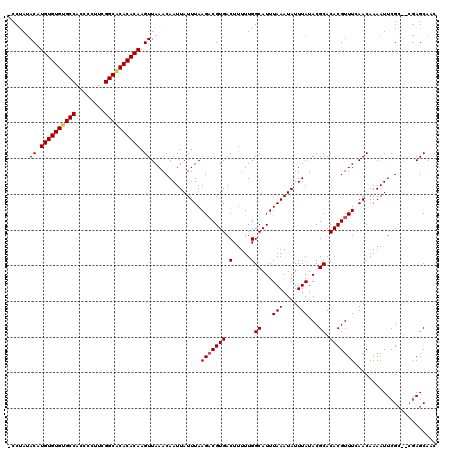
| Location | 528,888 – 528,988 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.63 |
| Mean single sequence MFE | -31.28 |
| Consensus MFE | -20.96 |
| Energy contribution | -20.80 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.67 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.533164 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 528888 100 + 22224390 UAUUUAAAUGCCAAAAAGUCACGUCUUAAAUAAUUGUUUAACUUGUGUGCGCCGAAGGGGUGGCACACACAUGUAUAGG----GGUGCGGG---------------ACGG-GCUUGCUAU ...............(((((.((((((..((..((((...((.((((((.((((......)))).)))))).)))))).----.))..)))---------------))))-))))..... ( -29.80) >DroSec_CAF1 15606 100 + 1 UAUUUAAAUGCCAAAAAGUCACGGCUUAAAUAAUUGUUUAACUUGUGUGUGCCGAAGGGGUGGCACACACAUGUAUAGG----CGUGCGGG---------------ACGG-GCUUGCUAU .......(((((...((((....)))).............((.(((((((((((......))))))))))).))...))----)))(((((---------------....-.)))))... ( -33.30) >DroSim_CAF1 15925 99 + 1 UAUUUAAAUGCCAAAAAGUCACGUCUUAAAUAAUUGUUUAACUUGUGUGUGCCGAAGGGGUGGCACACACAUGUAUAGG----GGUGCGGG---------------AGGG--GUUGCUAU ......(((.((.....(.(((.((((((((....)))))((.(((((((((((......))))))))))).))..)))----.))))...---------------.)).--)))..... ( -28.50) >DroEre_CAF1 20611 101 + 1 UAUUUAAAUGCCAAAAAGUCACGUCUUAAAUAAUUGUUUAACUUGUGUGUGCCGAAGGGGUGGCGCACACAUGUAUGGG----GGUGUGGG---------------GCGGAGGUUGCUAU ........((((.......(((.((((((((....)))))((.(((((((((((......))))))))))).))..)))----.)))...)---------------)))........... ( -29.20) >DroYak_CAF1 15033 120 + 1 UAUUUAAAUGCCAAAAAGUCACGUCUUAAAUAAUUGUUUAACUUGUGUGUGCCGAAGGGGUGGCACACACAUGUGUGGGUGUGGGUGUGGGUGUGUGUGAGGGGGUGCGGAGCUUGCUAU .......(((((....(.((((..(((((((....)))))((.(((((((((((......))))))))))).))..))..)))).)...))))).....((.((((.....)))).)).. ( -35.60) >consensus UAUUUAAAUGCCAAAAAGUCACGUCUUAAAUAAUUGUUUAACUUGUGUGUGCCGAAGGGGUGGCACACACAUGUAUAGG____GGUGCGGG_______________ACGG_GCUUGCUAU ..........((.....(.(((....(((((....)))))((.(((((((((((......))))))))))).))..........)))).))............................. (-20.96 = -20.80 + -0.16)

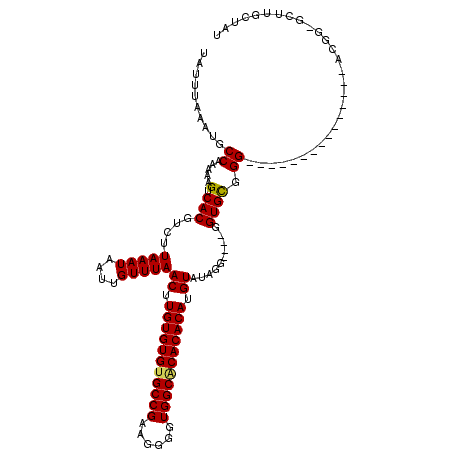
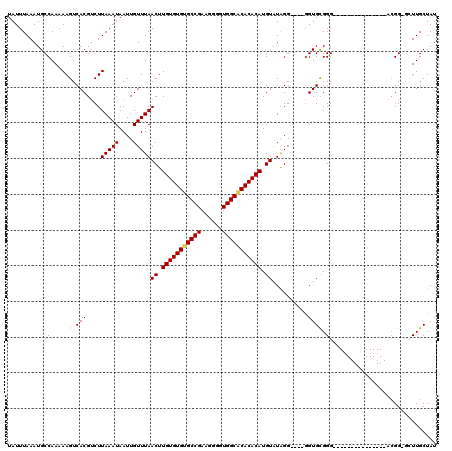
| Location | 528,888 – 528,988 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.63 |
| Mean single sequence MFE | -23.58 |
| Consensus MFE | -20.08 |
| Energy contribution | -19.92 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.04 |
| Structure conservation index | 0.85 |
| SVM decision value | 3.44 |
| SVM RNA-class probability | 0.999221 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 528888 100 - 22224390 AUAGCAAGC-CCGU---------------CCCGCACC----CCUAUACAUGUGUGUGCCACCCCUUCGGCGCACACAAGUUAAACAAUUAUUUAAGACGUGACUUUUUGGCAUUUAAAUA .......((-(.((---------------(.(((...----........((((((((((........))))))))))..(((((......)))))).)).))).....)))......... ( -25.00) >DroSec_CAF1 15606 100 - 1 AUAGCAAGC-CCGU---------------CCCGCACG----CCUAUACAUGUGUGUGCCACCCCUUCGGCACACACAAGUUAAACAAUUAUUUAAGCCGUGACUUUUUGGCAUUUAAAUA .......((-(...---------------....((((----.((..((.((((((((((........)))))))))).))((((......)))))).)))).......)))......... ( -27.94) >DroSim_CAF1 15925 99 - 1 AUAGCAAC--CCCU---------------CCCGCACC----CCUAUACAUGUGUGUGCCACCCCUUCGGCACACACAAGUUAAACAAUUAUUUAAGACGUGACUUUUUGGCAUUUAAAUA ...((...--....---------------..(((...----........((((((((((........))))))))))..(((((......)))))...)))........))......... ( -21.53) >DroEre_CAF1 20611 101 - 1 AUAGCAACCUCCGC---------------CCCACACC----CCCAUACAUGUGUGCGCCACCCCUUCGGCACACACAAGUUAAACAAUUAUUUAAGACGUGACUUUUUGGCAUUUAAAUA ............((---------------(.......----........((((((.(((........))).))))))(((((.....((....))....)))))....)))......... ( -19.40) >DroYak_CAF1 15033 120 - 1 AUAGCAAGCUCCGCACCCCCUCACACACACCCACACCCACACCCACACAUGUGUGUGCCACCCCUUCGGCACACACAAGUUAAACAAUUAUUUAAGACGUGACUUUUUGGCAUUUAAAUA ............((......((((.........................((((((((((........))))))))))..(((((......)))))...)))).......))......... ( -24.02) >consensus AUAGCAAGC_CCGU_______________CCCGCACC____CCUAUACAUGUGUGUGCCACCCCUUCGGCACACACAAGUUAAACAAUUAUUUAAGACGUGACUUUUUGGCAUUUAAAUA ...((..........................(((...............((((((((((........))))))))))..(((((......)))))...))).(.....)))......... (-20.08 = -19.92 + -0.16)

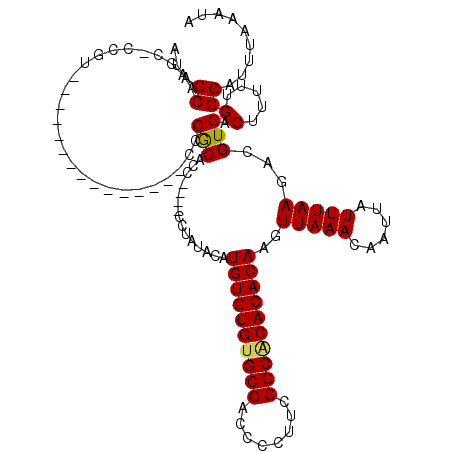
| Location | 528,928 – 529,028 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.00 |
| Mean single sequence MFE | -26.57 |
| Consensus MFE | -25.80 |
| Energy contribution | -26.08 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.97 |
| SVM decision value | 4.47 |
| SVM RNA-class probability | 0.999904 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 528928 100 - 22224390 GUAACAGCUGCAGUUAGCUGAAAAAGUUCUUAAAAUAGCAAUAGCAAGC-CCGU---------------CCCGCACC----CCUAUACAUGUGUGUGCCACCCCUUCGGCGCACACAAGU (((.((((((....)))))).................((....))....-....---------------........----....))).((((((((((........))))))))))... ( -27.10) >DroSec_CAF1 15646 100 - 1 GUAACAGCUGCAGUUAGCUGAAAAAGUUCUUAAAAUAGCAAUAGCAAGC-CCGU---------------CCCGCACG----CCUAUACAUGUGUGUGCCACCCCUUCGGCACACACAAGU (((.((((((....)))))).................((....))....-....---------------........----....))).((((((((((........))))))))))... ( -27.50) >DroSim_CAF1 15965 99 - 1 GUAACAGCUGCAGUUAGCUGAAAAAGUUCUUAAAAUAGCAAUAGCAAC--CCCU---------------CCCGCACC----CCUAUACAUGUGUGUGCCACCCCUUCGGCACACACAAGU (((.((((((....)))))).................((....))...--....---------------........----....))).((((((((((........))))))))))... ( -27.50) >DroEre_CAF1 20651 101 - 1 GUAACAGCUGCAGUUAGCUGAAAAAGUUCUUAAAAUAGCAAUAGCAACCUCCGC---------------CCCACACC----CCCAUACAUGUGUGCGCCACCCCUUCGGCACACACAAGU (((.((((((....)))))).................((....)).........---------------........----....))).((((((.(((........))).))))))... ( -23.60) >DroYak_CAF1 15073 120 - 1 GUAACAGCUGCAGUUAGCUGAAAAAGUUCUUAAAAUAGCAAUAGCAAGCUCCGCACCCCCUCACACACACCCACACCCACACCCACACAUGUGUGUGCCACCCCUUCGGCACACACAAGU ((...((((((.(((.((((...............))))))).)).))))..))................................((.((((((((((........)))))))))).)) ( -27.16) >consensus GUAACAGCUGCAGUUAGCUGAAAAAGUUCUUAAAAUAGCAAUAGCAAGC_CCGU_______________CCCGCACC____CCUAUACAUGUGUGUGCCACCCCUUCGGCACACACAAGU (((.((((((....)))))).................((....))........................................))).((((((((((........))))))))))... (-25.80 = -26.08 + 0.28)

| Location | 528,988 – 529,108 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -29.44 |
| Consensus MFE | -24.38 |
| Energy contribution | -25.18 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.559901 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 528988 120 + 22224390 UGCUAUUUUAAGAACUUUUUCAGCUAACUGCAGCUGUUACCGCCUGUAUGCUCCCUCUGGAGCCCCUGCAUGACCCUCAUUUCGCCAGCUACUUGCGUCAACAGGCACAAGGAUUCAUUA ...........(((((((..(((((......))))).....((((((..(((((....))))).......((((.............((.....))))))))))))..)))).))).... ( -27.91) >DroSec_CAF1 15706 116 + 1 UGCUAUUUUAAGAACUUUUUCAGCUAACUGCAGCUGUUACUGCCUGUACGCUCCCUCUGGAGCCCCUGCAUGGCCCUCAUUUCGCCAGCUACUUGCGUCAAC----ACAAGGAUUCAUUC .(((................(((((......))))).....(((((((.(((((....)))))...)))).)))............)))..((((.(....)----.))))......... ( -25.90) >DroSim_CAF1 16024 120 + 1 UGCUAUUUUAAGAACUUUUUCAGCUAACUGCAGCUGUUACUGCCUGUACGCUCCCUCUGGAGCCCCUGCAUGGCCCUCAUUUCGCCAGCUACUUGCGUCAACAGGCACAAGGAUUCAUUA ...........(((((((..(((((......)))))....(((((((..(((((....)))))....((.((((.........))))))...........))))))).)))).))).... ( -32.70) >DroEre_CAF1 20712 120 + 1 UGCUAUUUUAAGAACUUUUUCAGCUAACUGCAGCUGUUACUGCCUGUACGCUCCCUCUGGAGCCCCGGUAUGACCCCCACUUCGCCAGCUACUUGCGUCAACAGGCACAAGGAUUCAUUA ...........(((((((..(((((......)))))....(((((((((((.....((((......((........))......))))......))))..))))))).)))).))).... ( -29.50) >DroYak_CAF1 15153 120 + 1 UGCUAUUUUAAGAACUUUUUCAGCUAACUGCAGCUGUUACUGCCUGUACGCUCCCUCUGGAGCCCCUGCAGGACCCCCGUUUCGCCAGCUACUUGCGUCAACAGGCACAAGGAUUCAUUA ...........(((((((..(((((......)))))....(((((((..(((((....)))))....((..(((....)))..))..((.....))....))))))).)))).))).... ( -31.20) >consensus UGCUAUUUUAAGAACUUUUUCAGCUAACUGCAGCUGUUACUGCCUGUACGCUCCCUCUGGAGCCCCUGCAUGACCCUCAUUUCGCCAGCUACUUGCGUCAACAGGCACAAGGAUUCAUUA ...........(((((((..(((((......))))).....((((((..(((((....)))))...................(((.((...)).)))...))))))..)))).))).... (-24.38 = -25.18 + 0.80)

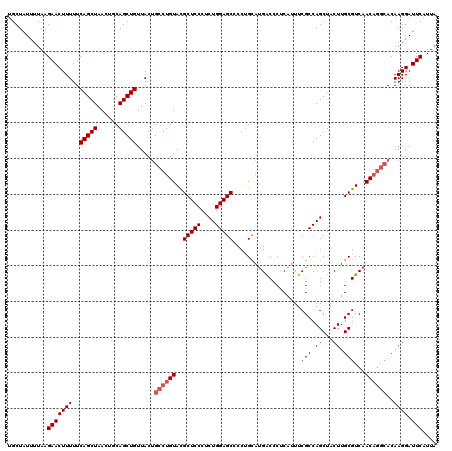
| Location | 528,988 – 529,108 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -37.26 |
| Consensus MFE | -31.82 |
| Energy contribution | -31.82 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.910280 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
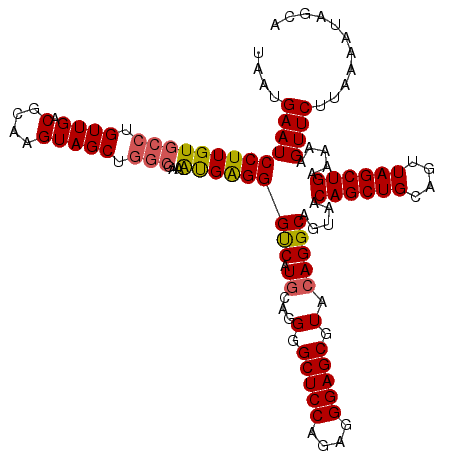
>X_DroMel_CAF1 528988 120 - 22224390 UAAUGAAUCCUUGUGCCUGUUGACGCAAGUAGCUGGCGAAAUGAGGGUCAUGCAGGGGCUCCAGAGGGAGCAUACAGGCGGUAACAGCUGCAGUUAGCUGAAAAAGUUCUUAAAAUAGCA ..((((.(((((.((((.((((.(....))))).))))....)))))))))((..(.(((((....)))))...)..)).....((((((....)))))).................... ( -37.60) >DroSec_CAF1 15706 116 - 1 GAAUGAAUCCUUGU----GUUGACGCAAGUAGCUGGCGAAAUGAGGGCCAUGCAGGGGCUCCAGAGGGAGCGUACAGGCAGUAACAGCUGCAGUUAGCUGAAAAAGUUCUUAAAAUAGCA .........(((((----(....))))))..((((......(((((((..(((..(.(((((....))))).)....)))....((((((....)))))).....)))))))...)))). ( -35.20) >DroSim_CAF1 16024 120 - 1 UAAUGAAUCCUUGUGCCUGUUGACGCAAGUAGCUGGCGAAAUGAGGGCCAUGCAGGGGCUCCAGAGGGAGCGUACAGGCAGUAACAGCUGCAGUUAGCUGAAAAAGUUCUUAAAAUAGCA ....((((.....(((((((..((....)).((((((.........)))).))....(((((....)))))..)))))))....((((((....)))))).....))))........... ( -37.30) >DroEre_CAF1 20712 120 - 1 UAAUGAAUCCUUGUGCCUGUUGACGCAAGUAGCUGGCGAAGUGGGGGUCAUACCGGGGCUCCAGAGGGAGCGUACAGGCAGUAACAGCUGCAGUUAGCUGAAAAAGUUCUUAAAAUAGCA ..((((..(((..((((.((((.(....))))).))))....)))..)))).((.(.(((((....))))).)...))......((((((....)))))).................... ( -37.00) >DroYak_CAF1 15153 120 - 1 UAAUGAAUCCUUGUGCCUGUUGACGCAAGUAGCUGGCGAAACGGGGGUCCUGCAGGGGCUCCAGAGGGAGCGUACAGGCAGUAACAGCUGCAGUUAGCUGAAAAAGUUCUUAAAAUAGCA ......(((((((((((.((((.(....))))).)))...)))))))).((((..(.(((((....))))).)....))))...((((((....)))))).................... ( -39.20) >consensus UAAUGAAUCCUUGUGCCUGUUGACGCAAGUAGCUGGCGAAAUGAGGGUCAUGCAGGGGCUCCAGAGGGAGCGUACAGGCAGUAACAGCUGCAGUUAGCUGAAAAAGUUCUUAAAAUAGCA ....(((((((((((((.((((.(....))))).)))...))))))(((.((...(.(((((....))))).).))))).....((((((....)))))).....))))........... (-31.82 = -31.82 + 0.00)

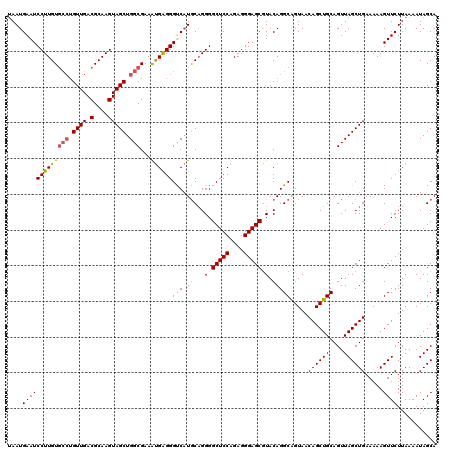
| Location | 529,028 – 529,148 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.50 |
| Mean single sequence MFE | -42.12 |
| Consensus MFE | -36.88 |
| Energy contribution | -37.52 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.801708 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 529028 120 - 22224390 UGUGCAUGAGUGUGGGCGGCUGAGUAAUUUGCGCUCGUAAUAAUGAAUCCUUGUGCCUGUUGACGCAAGUAGCUGGCGAAAUGAGGGUCAUGCAGGGGCUCCAGAGGGAGCAUACAGGCG (((..(((((((..((..((...))..))..))))))).)))((((.(((((.((((.((((.(....))))).))))....)))))))))((..(.(((((....)))))...)..)). ( -42.20) >DroSec_CAF1 15746 116 - 1 UGUGCAUGAGUGUGGGCGGCUGAGUAAUUUGUGCUCGUAAGAAUGAAUCCUUGU----GUUGACGCAAGUAGCUGGCGAAAUGAGGGCCAUGCAGGGGCUCCAGAGGGAGCGUACAGGCA ((((((((((((..((..((...))..))..)))))))..........((((((----((((.(....)))))((((.........)))).))))))(((((....)))))))))).... ( -37.70) >DroSim_CAF1 16064 120 - 1 UGUGCAUGAGUGUGGGCGGCUGAGUAAUUUGUGCUCGUAAUAAUGAAUCCUUGUGCCUGUUGACGCAAGUAGCUGGCGAAAUGAGGGCCAUGCAGGGGCUCCAGAGGGAGCGUACAGGCA (((((....(((((((((((.((....))...)).))).........(((((.((((.((((.(....))))).))))....)))))))))))....(((((....)))))))))).... ( -41.70) >DroEre_CAF1 20752 120 - 1 UGUGCAUGAGUGUGGGCGGCUGAGUAAUUUGCGCUCGUAAUAAUGAAUCCUUGUGCCUGUUGACGCAAGUAGCUGGCGAAGUGGGGGUCAUACCGGGGCUCCAGAGGGAGCGUACAGGCA ((((((((((((..((..((...))..))..)))))))....((((..(((..((((.((((.(....))))).))))....)))..))))......(((((....)))))))))).... ( -44.30) >DroYak_CAF1 15193 120 - 1 UGUGCAUGAGUGUGGGCGGCUGAGUAAUUUGCGCUCGUAAUAAUGAAUCCUUGUGCCUGUUGACGCAAGUAGCUGGCGAAACGGGGGUCCUGCAGGGGCUCCAGAGGGAGCGUACAGGCA ((((((((((((..((..((...))..))..))))))).........(((((.((((.((((.(....))))).)))......(((((((.....)))))))).)))))..))))).... ( -44.70) >consensus UGUGCAUGAGUGUGGGCGGCUGAGUAAUUUGCGCUCGUAAUAAUGAAUCCUUGUGCCUGUUGACGCAAGUAGCUGGCGAAAUGAGGGUCAUGCAGGGGCUCCAGAGGGAGCGUACAGGCA ((((((((((((..((..((...))..))..)))))))..........(((((((((.((((.(....))))).)))...(((.....)))))))))(((((....)))))))))).... (-36.88 = -37.52 + 0.64)

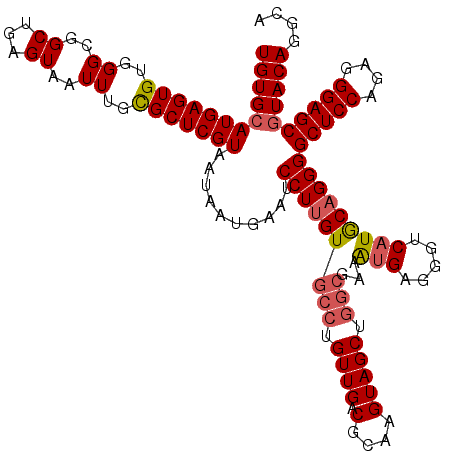
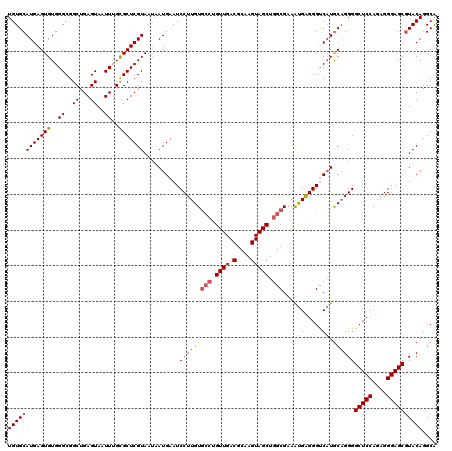
| Location | 529,068 – 529,188 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.83 |
| Mean single sequence MFE | -38.68 |
| Consensus MFE | -36.20 |
| Energy contribution | -36.80 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.54 |
| SVM RNA-class probability | 0.995084 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 529068 120 + 22224390 UUCGCCAGCUACUUGCGUCAACAGGCACAAGGAUUCAUUAUUACGAGCGCAAAUUACUCAGCCGCCCACACUCAUGCACAAUUAAGUAAUGCGUGUGUGAGGGUGGUGGUUUGCAUGUGU ..(((..(((.((((.(((....))).))))(((.....)))...)))(((((((.....(((((((.(((.((((((...........)))))).))).))))))))))))))..))). ( -40.20) >DroSec_CAF1 15786 116 + 1 UUCGCCAGCUACUUGCGUCAAC----ACAAGGAUUCAUUCUUACGAGCACAAAUUACUCAGCCGCCCACACUCAUGCACAAUUAAGUAAUGCGUGUGUGAGGGUGGUGGUUUGCAUGUGU ...((.((((.((((.(....)----.)))).............(((.........))).(((((((.(((.((((((...........)))))).))).))))))))))).))...... ( -34.60) >DroSim_CAF1 16104 120 + 1 UUCGCCAGCUACUUGCGUCAACAGGCACAAGGAUUCAUUAUUACGAGCACAAAUUACUCAGCCGCCCACACUCAUGCACAAUUAAGUAAUGCGUGUGUGAGGGUGGUGGUUUGCAUGUGU ...((.((((.((((.(((....))).)))).............(((.........))).(((((((.(((.((((((...........)))))).))).))))))))))).))...... ( -38.20) >DroEre_CAF1 20792 120 + 1 UUCGCCAGCUACUUGCGUCAACAGGCACAAGGAUUCAUUAUUACGAGCGCAAAUUACUCAGCCGCCCACACUCAUGCACAAUUAAGUAAUGCGUGUGUGAGGGUGGUGGUUUGCAUGUGU ..(((..(((.((((.(((....))).))))(((.....)))...)))(((((((.....(((((((.(((.((((((...........)))))).))).))))))))))))))..))). ( -40.20) >DroYak_CAF1 15233 120 + 1 UUCGCCAGCUACUUGCGUCAACAGGCACAAGGAUUCAUUAUUACGAGCGCAAAUUACUCAGCCGCCCACACUCAUGCACAAUUAAGUAAUGCGUGUGUGAGGGUGGUGGUUUGCAUGUGU ..(((..(((.((((.(((....))).))))(((.....)))...)))(((((((.....(((((((.(((.((((((...........)))))).))).))))))))))))))..))). ( -40.20) >consensus UUCGCCAGCUACUUGCGUCAACAGGCACAAGGAUUCAUUAUUACGAGCGCAAAUUACUCAGCCGCCCACACUCAUGCACAAUUAAGUAAUGCGUGUGUGAGGGUGGUGGUUUGCAUGUGU ...((.((((.((((.(((....))).)))).............(((.........))).(((((((.(((.((((((...........)))))).))).))))))))))).))...... (-36.20 = -36.80 + 0.60)

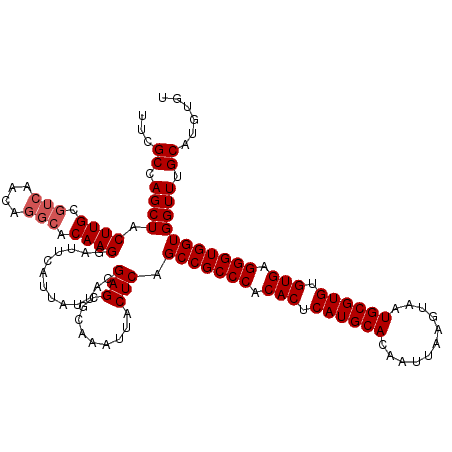
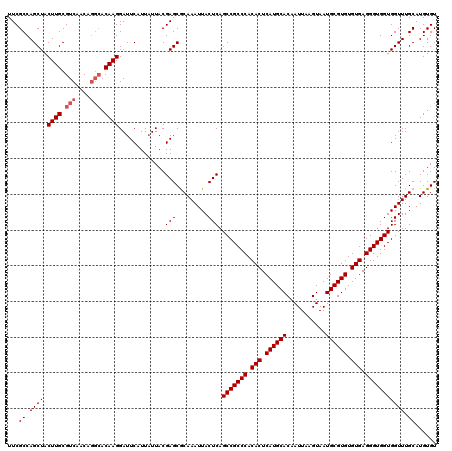
| Location | 529,108 – 529,228 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.50 |
| Mean single sequence MFE | -36.45 |
| Consensus MFE | -36.69 |
| Energy contribution | -36.45 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.35 |
| Structure conservation index | 1.01 |
| SVM decision value | 3.25 |
| SVM RNA-class probability | 0.998846 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 529108 120 + 22224390 UUACGAGCGCAAAUUACUCAGCCGCCCACACUCAUGCACAAUUAAGUAAUGCGUGUGUGAGGGUGGUGGUUUGCAUGUGUGCUGCUGUUAAUUUUCAUAUUAAAUCCGUUAAAAUGUGCA ......(((((.........(((((((.(((.((((((...........)))))).))).)))))))((((((.(((((................)))))))))))........))))). ( -36.29) >DroSec_CAF1 15822 120 + 1 UUACGAGCACAAAUUACUCAGCCGCCCACACUCAUGCACAAUUAAGUAAUGCGUGUGUGAGGGUGGUGGUUUGCAUGUGUGCUGCUGUUAAUUUUCAUAUUAAAUCCGUUAAAAUGUGCA ......(((((.........(((((((.(((.((((((...........)))))).))).)))))))((((((.(((((................)))))))))))........))))). ( -36.69) >DroSim_CAF1 16144 120 + 1 UUACGAGCACAAAUUACUCAGCCGCCCACACUCAUGCACAAUUAAGUAAUGCGUGUGUGAGGGUGGUGGUUUGCAUGUGUGCUGCUGUUAAUUUUCAUAUUAAAUCCGUUAAAAUGUGCA ......(((((.........(((((((.(((.((((((...........)))))).))).)))))))((((((.(((((................)))))))))))........))))). ( -36.69) >DroEre_CAF1 20832 120 + 1 UUACGAGCGCAAAUUACUCAGCCGCCCACACUCAUGCACAAUUAAGUAAUGCGUGUGUGAGGGUGGUGGUUUGCAUGUGUGCUGCUGUUAAUUUUCAUAUUAAAUCCGUUAAAAUGUGCA ......(((((.........(((((((.(((.((((((...........)))))).))).)))))))((((((.(((((................)))))))))))........))))). ( -36.29) >DroYak_CAF1 15273 120 + 1 UUACGAGCGCAAAUUACUCAGCCGCCCACACUCAUGCACAAUUAAGUAAUGCGUGUGUGAGGGUGGUGGUUUGCAUGUGUGCUGCUGUUAAUUUUCAUAUUAAAUCCGUUAAAAUGUGCA ......(((((.........(((((((.(((.((((((...........)))))).))).)))))))((((((.(((((................)))))))))))........))))). ( -36.29) >consensus UUACGAGCGCAAAUUACUCAGCCGCCCACACUCAUGCACAAUUAAGUAAUGCGUGUGUGAGGGUGGUGGUUUGCAUGUGUGCUGCUGUUAAUUUUCAUAUUAAAUCCGUUAAAAUGUGCA ......(((((.........(((((((.(((.((((((...........)))))).))).)))))))((((((.(((((................)))))))))))........))))). (-36.69 = -36.45 + -0.24)

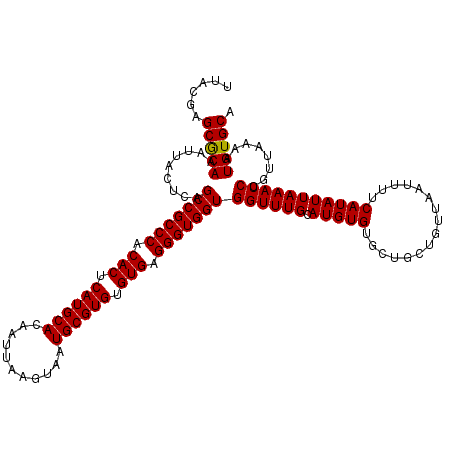
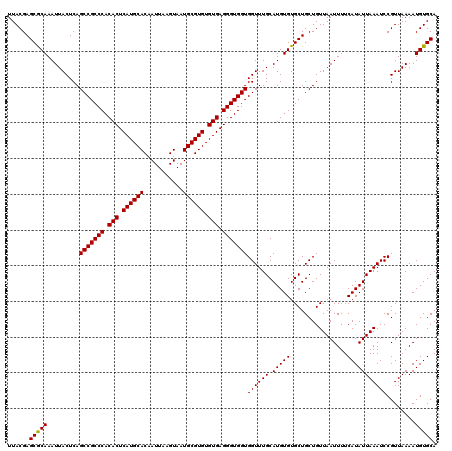
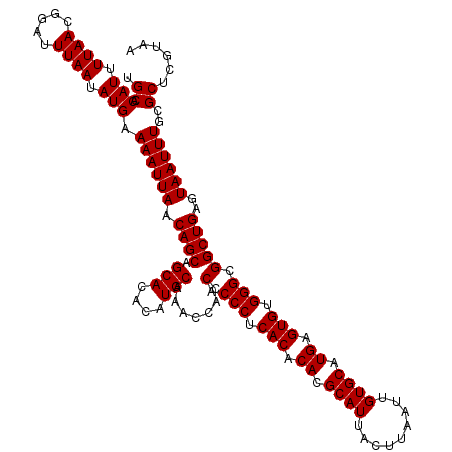
| Location | 529,108 – 529,228 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.50 |
| Mean single sequence MFE | -29.38 |
| Consensus MFE | -27.78 |
| Energy contribution | -27.78 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.882481 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 529108 120 - 22224390 UGCACAUUUUAACGGAUUUAAUAUGAAAAUUAACAGCAGCACACAUGCAAACCACCACCCUCACACACGCAUUACUUAAUUGUGCAUGAGUGUGGGCGGCUGAGUAAUUUGCGCUCGUAA .((.(((.((((.....)))).))).((((((.((((.(((....))).......(.(((.(((.((.((((.........)))).)).))).))).)))))..))))))))........ ( -28.10) >DroSec_CAF1 15822 120 - 1 UGCACAUUUUAACGGAUUUAAUAUGAAAAUUAACAGCAGCACACAUGCAAACCACCACCCUCACACACGCAUUACUUAAUUGUGCAUGAGUGUGGGCGGCUGAGUAAUUUGUGCUCGUAA .(((((..(((......(((((......)))))((((.(((....))).......(.(((.(((.((.((((.........)))).)).))).))).)))))..)))..)))))...... ( -31.30) >DroSim_CAF1 16144 120 - 1 UGCACAUUUUAACGGAUUUAAUAUGAAAAUUAACAGCAGCACACAUGCAAACCACCACCCUCACACACGCAUUACUUAAUUGUGCAUGAGUGUGGGCGGCUGAGUAAUUUGUGCUCGUAA .(((((..(((......(((((......)))))((((.(((....))).......(.(((.(((.((.((((.........)))).)).))).))).)))))..)))..)))))...... ( -31.30) >DroEre_CAF1 20832 120 - 1 UGCACAUUUUAACGGAUUUAAUAUGAAAAUUAACAGCAGCACACAUGCAAACCACCACCCUCACACACGCAUUACUUAAUUGUGCAUGAGUGUGGGCGGCUGAGUAAUUUGCGCUCGUAA .((.(((.((((.....)))).))).((((((.((((.(((....))).......(.(((.(((.((.((((.........)))).)).))).))).)))))..))))))))........ ( -28.10) >DroYak_CAF1 15273 120 - 1 UGCACAUUUUAACGGAUUUAAUAUGAAAAUUAACAGCAGCACACAUGCAAACCACCACCCUCACACACGCAUUACUUAAUUGUGCAUGAGUGUGGGCGGCUGAGUAAUUUGCGCUCGUAA .((.(((.((((.....)))).))).((((((.((((.(((....))).......(.(((.(((.((.((((.........)))).)).))).))).)))))..))))))))........ ( -28.10) >consensus UGCACAUUUUAACGGAUUUAAUAUGAAAAUUAACAGCAGCACACAUGCAAACCACCACCCUCACACACGCAUUACUUAAUUGUGCAUGAGUGUGGGCGGCUGAGUAAUUUGCGCUCGUAA .((.(((.((((.....)))).))).((((((.((((.(((....))).......(.(((.(((.((.((((.........)))).)).))).))).)))))..))))))..))...... (-27.78 = -27.78 + -0.00)

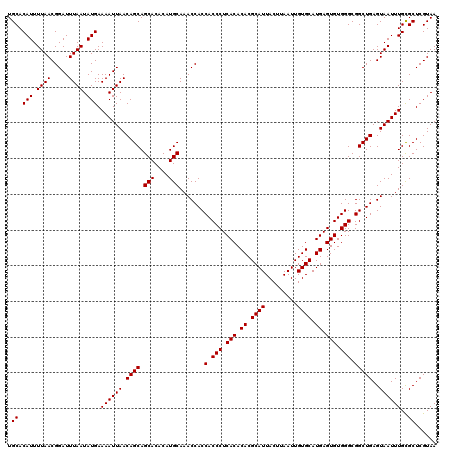
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:36:25 2006