
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,301,565 – 4,301,748 |
| Length | 183 |
| Max. P | 0.736735 |

| Location | 4,301,565 – 4,301,671 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.39 |
| Mean single sequence MFE | -30.55 |
| Consensus MFE | -19.04 |
| Energy contribution | -19.44 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.62 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.510962 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4301565 106 + 22224390 CCUGGCUAACCC----------CGCUAAAUCCCUUAACCCUUAACU----ACGAUGGGCUUGGCCAGAGUGAGAUUUCGAUUUGGCCUGGCAUUUUUUGGGCUUAUCUACAUCUGCGCUU .((((((((.((----------(((.....................----..).)))).))))))))((((((((...(((..((((..(......)..)))).)))...)))).)))). ( -31.30) >DroSec_CAF1 87998 113 + 1 CCUGGCUAACCCCGUU-AACCCCGCUAAAUCCCUUAACCCUU-ACU----GCGAUGGGCUUGGCCAGAGUGAGAUUUCGAUUUGGCCUGGCAUUU-UUGGGCUUAUCUACAUCUCUGCUU .((((((((.(((((.-.....(((.................-...----)))))))).))))))))((((((((...(((..((((..(.....-)..)))).)))...))))).))). ( -35.45) >DroSim_CAF1 81530 114 + 1 CCUGGCUAACCCCGCUGAACCCCGCCAAAUCCCUUAACCCUU-ACU----GCGAUGGGCUUGGCCAGAGUGAGAUUUCGAUUUGGCCUGGCAUUU-UUGGGCUUAUCUACAUCUCUGCUU .((((((((((((((...........................-...----)))..))).))))))))((((((((...(((..((((..(.....-)..)))).)))...))))).))). ( -33.81) >DroEre_CAF1 94982 103 + 1 CCCCUCCA---------AGCCCCUCUAAGCCCCUUAACCCUU-ACU----GCGAUGGGCUUGGCCAGAGUGAGAUUUCGAUUUGGCCUGGCAUUU-UAGCGCUUAUCUACAUC--UGCUU ......((---------(((((.((((((..........)))-)..----..)).)))))))(((((.((.((((....)))).)))))))....-....((...........--.)).. ( -25.50) >DroYak_CAF1 89711 98 + 1 CUC-------------------CGCUAAAUCCCUUAACCCUU-GCUGCCUGCGAUGGGCUUGGCCAGAAUGAGAUUUCGAUUUGGCCUGGCAUUU-UUGGGCUUAUCUACAUCU-UGCUU ...-------------------........(((....((.((-((.....)))).))(((.(((((((.((......)).))))))).)))....-..))).............-..... ( -26.70) >consensus CCUGGCUAACCC_____A_CCCCGCUAAAUCCCUUAACCCUU_ACU____GCGAUGGGCUUGGCCAGAGUGAGAUUUCGAUUUGGCCUGGCAUUU_UUGGGCUUAUCUACAUCU_UGCUU ....................................................(((((((((.(((((.((.((((....)))).))))))).......)))))))))............. (-19.04 = -19.44 + 0.40)



| Location | 4,301,595 – 4,301,711 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.81 |
| Mean single sequence MFE | -36.36 |
| Consensus MFE | -31.26 |
| Energy contribution | -31.22 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.736735 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4301595 116 + 22224390 UUAACU----ACGAUGGGCUUGGCCAGAGUGAGAUUUCGAUUUGGCCUGGCAUUUUUUGGGCUUAUCUACAUCUGCGCUUGUUCGCAUUUGUGGAAAACACUUGGCACAGAUAAGAGGCA ....((----(.((...(((.(((((((.((......)).))))))).)))...)).)))((((.((((((..((((......))))..)))))).....((((.......)))))))). ( -34.80) >DroSec_CAF1 88037 114 + 1 UU-ACU----GCGAUGGGCUUGGCCAGAGUGAGAUUUCGAUUUGGCCUGGCAUUU-UUGGGCUUAUCUACAUCUCUGCUUGUCCGCAUUUGUGGAAAACACUUGGCACAGAUAAGAGGCA ..-...----(((((((((((.(((((.((.((((....)))).)))))))....-..)))))))))...((((.((((..(((((....)))))........)))).)))).....)). ( -36.40) >DroSim_CAF1 81570 114 + 1 UU-ACU----GCGAUGGGCUUGGCCAGAGUGAGAUUUCGAUUUGGCCUGGCAUUU-UUGGGCUUAUCUACAUCUCUGCUUGUCCGCAUUUGUGGAAAACACUUGGCACAGAUAAGAGGCA ..-...----(((((((((((.(((((.((.((((....)))).)))))))....-..)))))))))...((((.((((..(((((....)))))........)))).)))).....)). ( -36.40) >DroEre_CAF1 95013 112 + 1 UU-ACU----GCGAUGGGCUUGGCCAGAGUGAGAUUUCGAUUUGGCCUGGCAUUU-UAGCGCUUAUCUACAUC--UGCUUGUCCGCAUUUGUGGAAAACACUUGGCACAGAUAAGGGGCA ..-...----((.....(((.(((((((.((......)).))))))).)))....-...(.(((((((.((..--((....(((((....)))))...))..))....))))))).))). ( -35.20) >DroYak_CAF1 89732 117 + 1 UU-GCUGCCUGCGAUGGGCUUGGCCAGAAUGAGAUUUCGAUUUGGCCUGGCAUUU-UUGGGCUUAUCUACAUCU-UGCUUGUCCGCAUUUGUGGAAAACACUUGGCGCAGAUAAGAGGCA ..-...((((..((((((((.(((((((.((......)).)))))))...((...-.)))))))))).......-..(((((((((....(((.....)))...)))..)))))))))). ( -39.00) >consensus UU_ACU____GCGAUGGGCUUGGCCAGAGUGAGAUUUCGAUUUGGCCUGGCAUUU_UUGGGCUUAUCUACAUCU_UGCUUGUCCGCAUUUGUGGAAAACACUUGGCACAGAUAAGAGGCA .................(((.(((((((.((......)).))))))).))).........(((..(((..((((.((((..(((((....)))))........)))).)))).)))))). (-31.26 = -31.22 + -0.04)

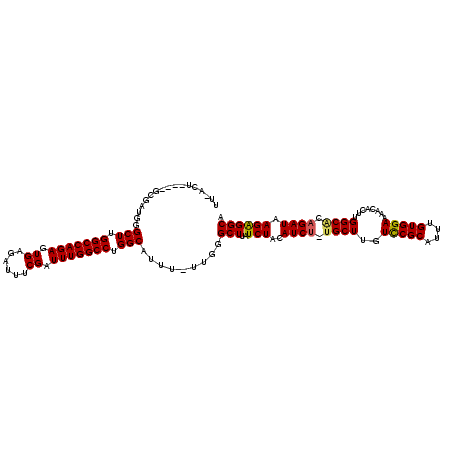
| Location | 4,301,631 – 4,301,748 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.65 |
| Mean single sequence MFE | -32.42 |
| Consensus MFE | -24.52 |
| Energy contribution | -24.40 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.621581 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4301631 117 - 22224390 UCCUCUCAGUCCUGGG---UGCAGGCUUUCAAAAUUUUCCUGCCUCUUAUCUGUGCCAAGUGUUUUCCACAAAUGCGAACAAGCGCAGAUGUAGAUAAGCCCAAAAAAUGCCAGGCCAAA .(((..((....((((---((((((.............))))).((((((((((((....(((((..((....)).))))).))))))))).)))...))))).....))..)))..... ( -31.12) >DroSec_CAF1 88072 116 - 1 UCCUCGCAGUUCUCGG---GGCAGGCUUUCAAAAUUUUCCUGCCUCUUAUCUGUGCCAAGUGUUUUCCACAAAUGCGGACAAGCAGAGAUGUAGAUAAGCCCAA-AAAUGCCAGGCCAAA .(((.(((.......(---(((.(((...............)))((((((((.(((....((...(((.(....).))))).))).))))).)))...))))..-...))).)))..... ( -31.06) >DroSim_CAF1 81605 116 - 1 UCCUCGCAGCUCUGGG---GGCAGGCUUUCAAAAUUUUCCUGCCUCUUAUCUGUGCCAAGUGUUUUCCACAAAUGCGGACAAGCAGAGAUGUAGAUAAGCCCAA-AAAUGCCAGGCCAAA .(((.(((....((((---((((((.............))))))((((((((.(((....((...(((.(....).))))).))).))))).)))....)))).-...))).)))..... ( -34.92) >DroEre_CAF1 95048 113 - 1 UCCUC-CAGCCCUGGG---GGCAGACUUUCAAAAUUUUCCUGCCCCUUAUCUGUGCCAAGUGUUUUCCACAAAUGCGGACAAGCA--GAUGUAGAUAAGCGCUA-AAAUGCCAGGCCAAA .....-..(.((((((---(((((...............)))))))............(((((((((.(((..(((......)))--..))).)).))))))).-......)))).)... ( -33.26) >DroYak_CAF1 89771 117 - 1 UUCUC-CAACCUUGGGGAGGGCAGACUUUCAAAAUUUUCCUGCCUCUUAUCUGCGCCAAGUGUUUUCCACAAAUGCGGACAAGCA-AGAUGUAGAUAAGCCCAA-AAAUGCCAGGCCAAA .....-.....(((((..((((((...............))))))((((((((((.(...((((((((.(....).))).)))))-.).)))))))))))))))-............... ( -31.76) >consensus UCCUC_CAGCCCUGGG___GGCAGGCUUUCAAAAUUUUCCUGCCUCUUAUCUGUGCCAAGUGUUUUCCACAAAUGCGGACAAGCA_AGAUGUAGAUAAGCCCAA_AAAUGCCAGGCCAAA ..........(((((....(((((...............))))).(((((((((..(...((((((((.(....).))).)))))..)..)))))))))...........)))))..... (-24.52 = -24.40 + -0.12)

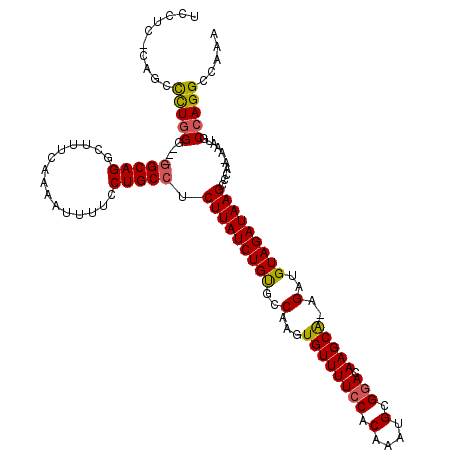
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:09:09 2006