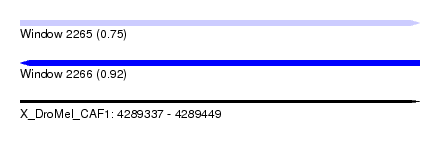
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,289,337 – 4,289,449 |
| Length | 112 |
| Max. P | 0.917412 |

| Location | 4,289,337 – 4,289,449 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 79.27 |
| Mean single sequence MFE | -29.03 |
| Consensus MFE | -12.34 |
| Energy contribution | -12.14 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.43 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.748358 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4289337 112 + 22224390 AGAACCUAAUUGAGGUUGAACCAAUUAGUGGCG------GGUCGCCGGGGAUCUUUGCCCC-ACAAAAAAAGAGCCAAGGCAAAUAGAUAGGCAAACAAGAGCCUACAACAAGUUCGCC .....(((((((.(......)))))))).((((------..(.(((..((.(((((.....-......))))).))..))).......(((((........)))))......)..)))) ( -32.30) >DroSec_CAF1 75755 94 + 1 AGAACCUAAUUGAGGUUGAACCAAUUAGUGGCG------GGUCGCCGGGGAUCUUUGCCCC-A----AAAAGAGCUAAGGC------------AAACAAGAGCCUACAAUA--UUCGCC .....(((((((.(......)))))))).((((------(((....((((.......))))-.----..........((((------------........)))).....)--)))))) ( -26.00) >DroSim_CAF1 70060 94 + 1 AGAACCUAAUUGAGGUUGAACCAAUUAGUGGCG------GGUCGCCGGGGAUCUUUGCCCC-A----AAAAGAGCCAAGGC------------AAACAAGAGCCUACAAUA--UUCGCC .....(((((((.(......)))))))).((((------(.((...((((.......))))-.----....)).)).((((------------........))))......--...))) ( -26.90) >DroEre_CAF1 82925 107 + 1 AGAACCUAAUUGAGGUUGAACCAAUUAGUGGCG------GGUCGCCAGGGAUCUUUGGCCCGA----AAAAAGGCCAAAGAAAAUAGGUAUGCAAGUAAGAGCCUACAAAA--GUCGCC .....(((((((.(......))))))))(((((------...)))))(((((((((((((...----.....)))))))))......(((.((........)).)))....--.)).)) ( -30.70) >DroYak_CAF1 77265 112 + 1 AGAACCUAAUUGAGAUUAAACCAAUUAGUGGGAUCUUAAAUUAGCCAGGGAUCUUUGACCC-A----AAAAGGGUCAAGGAAAAUAGGUAUAUAAACAAGAGCCUACAAUA--UGCGUC ......((((((.(......)))))))(((((.((((......(((.....((((((((((-.----....)))))))))).....)))........)))).)))))....--...... ( -29.24) >consensus AGAACCUAAUUGAGGUUGAACCAAUUAGUGGCG______GGUCGCCGGGGAUCUUUGCCCC_A____AAAAGAGCCAAGGCAAAUAG_UA___AAACAAGAGCCUACAAUA__UUCGCC .....(((((((.(......))))))))(((((.........))))).((.(((((............))))).))........................................... (-12.34 = -12.14 + -0.20)



| Location | 4,289,337 – 4,289,449 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 79.27 |
| Mean single sequence MFE | -30.09 |
| Consensus MFE | -15.74 |
| Energy contribution | -16.82 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.52 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.917412 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4289337 112 - 22224390 GGCGAACUUGUUGUAGGCUCUUGUUUGCCUAUCUAUUUGCCUUGGCUCUUUUUUUGU-GGGGCAAAGAUCCCCGGCGACC------CGCCACUAAUUGGUUCAACCUCAAUUAGGUUCU ((((........((((((........))))))....(((((...((.........))-((((.......)))))))))..------)))).((((((((.......))))))))..... ( -32.80) >DroSec_CAF1 75755 94 - 1 GGCGAA--UAUUGUAGGCUCUUGUUU------------GCCUUAGCUCUUUU----U-GGGGCAAAGAUCCCCGGCGACC------CGCCACUAAUUGGUUCAACCUCAAUUAGGUUCU ((((..--..((((.((....(.(((------------(((((((......)----)-)))))))).)...)).))))..------)))).((((((((.......))))))))..... ( -27.90) >DroSim_CAF1 70060 94 - 1 GGCGAA--UAUUGUAGGCUCUUGUUU------------GCCUUGGCUCUUUU----U-GGGGCAAAGAUCCCCGGCGACC------CGCCACUAAUUGGUUCAACCUCAAUUAGGUUCU ((((..--..((((.((....(.(((------------((((..(......)----.-.))))))).)...)).))))..------)))).((((((((.......))))))))..... ( -28.70) >DroEre_CAF1 82925 107 - 1 GGCGAC--UUUUGUAGGCUCUUACUUGCAUACCUAUUUUCUUUGGCCUUUUU----UCGGGCCAAAGAUCCCUGGCGACC------CGCCACUAAUUGGUUCAACCUCAAUUAGGUUCU ((((..--...((((((......))))))..((.....((((((((((....----..)))))))))).....)).....------)))).((((((((.......))))))))..... ( -35.60) >DroYak_CAF1 77265 112 - 1 GACGCA--UAUUGUAGGCUCUUGUUUAUAUACCUAUUUUCCUUGACCCUUUU----U-GGGUCAAAGAUCCCUGGCUAAUUUAAGAUCCCACUAAUUGGUUUAAUCUCAAUUAGGUUCU (((...--.......((.(((((........((.....((.(((((((....----.-))))))).)).....))......))))).))..((((((((.......))))))))))).. ( -25.44) >consensus GGCGAA__UAUUGUAGGCUCUUGUUU___UA_CUAUUUGCCUUGGCUCUUUU____U_GGGGCAAAGAUCCCCGGCGACC______CGCCACUAAUUGGUUCAACCUCAAUUAGGUUCU ((((........((((((....))))))..............................((((.......)))).............)))).((((((((.......))))))))..... (-15.74 = -16.82 + 1.08)

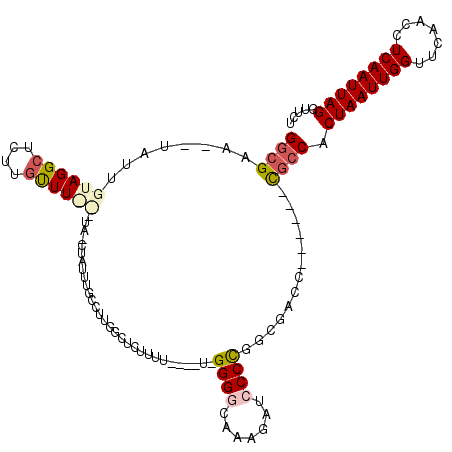

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:09:01 2006