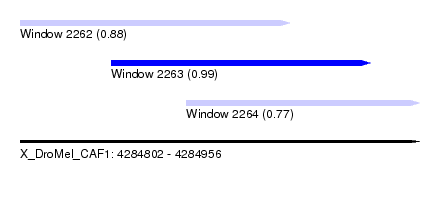
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,284,802 – 4,284,956 |
| Length | 154 |
| Max. P | 0.993438 |

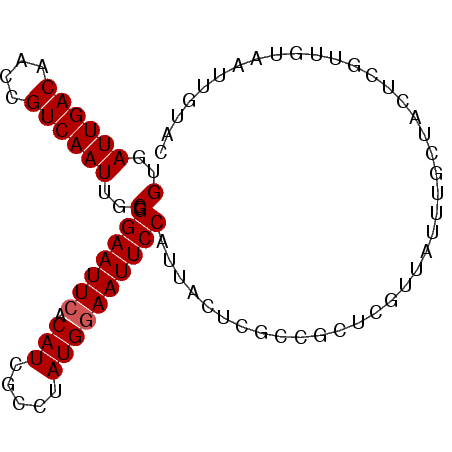
| Location | 4,284,802 – 4,284,906 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 95.89 |
| Mean single sequence MFE | -30.20 |
| Consensus MFE | -26.93 |
| Energy contribution | -26.71 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.880656 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4284802 104 + 22224390 AGGGGA--AAACAGAAAGUGGAACAAGUUAGGUGACAGGCAGAAACAACUUGGCCAGAUUCAAACUGUGAUUGACAACCGUCAAUUGCGGGAAUUCACAUCGCCUA (((.((--.....(((..(((..((((((.(....).(.......))))))).)))..)))...(((..((((((....))))))..))).........)).))). ( -30.50) >DroEre_CAF1 78372 104 + 1 AGGGGA--UAACAGAAAGUGGAACAAGUUAGGUGACAGGCAGAAACAACUUGGCCAGAUCCAAGCUGUGAUUGACAACCGUCAAUUGCGGGAAUUCACAUCGCCAA .((.((--(.........((((.(......(....).(((.((......)).))).).))))..(((..((((((....))))))..)))........))).)).. ( -29.90) >DroYak_CAF1 72845 106 + 1 AGGGGAUAUAACAGAAAGUGGAACAAGUUAGGUGACAGGCAGAAACAACUUGGCCAGAUCCAAACUGUGAUUGACAACCGUCAAUUGCGGGAAUUCACAUCGCCGA .((.(((...........((((.(......(....).(((.((......)).))).).))))..(((..((((((....))))))..)))........))).)).. ( -30.20) >consensus AGGGGA__UAACAGAAAGUGGAACAAGUUAGGUGACAGGCAGAAACAACUUGGCCAGAUCCAAACUGUGAUUGACAACCGUCAAUUGCGGGAAUUCACAUCGCCAA .((.((............((((.(..(((((((..............)))))))..).))))..(((..((((((....))))))..))).........)).)).. (-26.93 = -26.71 + -0.22)

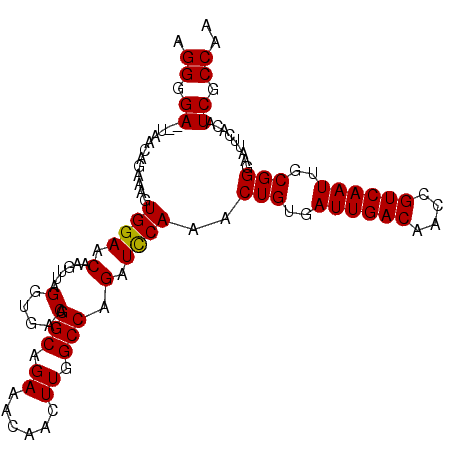

| Location | 4,284,837 – 4,284,937 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 96.30 |
| Mean single sequence MFE | -28.00 |
| Consensus MFE | -25.98 |
| Energy contribution | -26.02 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.40 |
| SVM RNA-class probability | 0.993438 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4284837 100 + 22224390 GGCAGAAACAACUUGGCCAGAUUCAAACUGUGAUUGACAACCGUCAAUUGCGGGAAUUCACAUCGCCUAUGGAAUUCCAUUACUCGCCGCUCGUUAUUUG ..(((((((....((((.((........((..((((((....))))))..))(((((((.(((.....))))))))))....)).))))...))).)))) ( -27.20) >DroSec_CAF1 71341 100 + 1 GGCAGAAACAACUUGGCCAGAUCCAAACUGUGAUUGACAACCGUCAAUUGCGGGAAUUCACAUCGCCUAUGGAAUUCCAUUUCUCGCCGCUCGUUAUUUG ((((((((....((((......))))..((..((((((....))))))..))(((((((.(((.....)))))))))).))))).)))............ ( -30.20) >DroSim_CAF1 65616 100 + 1 GGCAGAAACAACUUGGCCAGAUCCAAACUGUGAUUGACAACCGUCAAUUGCGGGAAUUCACAUCGCCUAUGGAAUUCCAUUACUCGCCGCUCGUUAUUUG (((((.......((((......))))..((..((((((....))))))..))(((((((.(((.....))))))))))....)).)))............ ( -27.30) >DroEre_CAF1 78407 100 + 1 GGCAGAAACAACUUGGCCAGAUCCAAGCUGUGAUUGACAACCGUCAAUUGCGGGAAUUCACAUCGCCAAUGAAAUUCCAUUACUCGCCGCUCGUUCUUCA (((.(((....(((((......)))))(((..((((((....))))))..)))...)))........((((......))))....)))............ ( -25.60) >DroYak_CAF1 72882 100 + 1 GGCAGAAACAACUUGGCCAGAUCCAAACUGUGAUUGACAACCGUCAAUUGCGGGAAUUCACAUCGCCGAUGGAAUUCCAUUACUCGCCGCUCGUUAUUCG ....(((..(((.((((.((........((..((((((....))))))..))(((((((.((((...)))))))))))....)).))))...))).))). ( -29.70) >consensus GGCAGAAACAACUUGGCCAGAUCCAAACUGUGAUUGACAACCGUCAAUUGCGGGAAUUCACAUCGCCUAUGGAAUUCCAUUACUCGCCGCUCGUUAUUUG (((.........((((......))))..((..((((((....))))))..))(((((((.(((.....)))))))))).......)))............ (-25.98 = -26.02 + 0.04)

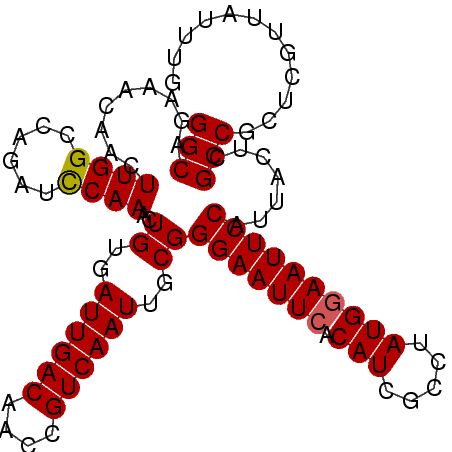

| Location | 4,284,866 – 4,284,956 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 94.33 |
| Mean single sequence MFE | -20.46 |
| Consensus MFE | -18.94 |
| Energy contribution | -19.14 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.772376 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4284866 90 + 22224390 GUGAUUGACAACCGUCAAUUGCGGGAAUUCACAUCGCCUAUGGAAUUCCAUUACUCGCCGCUCGUUAUUUGCUACUCGUUGUUAUUGUAC (..((((((....))))))..).(((((((.(((.....))))))))))..(((..((.((..((........))..)).))....))). ( -20.60) >DroSec_CAF1 71370 90 + 1 GUGAUUGACAACCGUCAAUUGCGGGAAUUCACAUCGCCUAUGGAAUUCCAUUUCUCGCCGCUCGUUAUUUGCUACUCGUUGUAAUUGUAC (..((((((....))))))..).(((((((.(((.....))))))))))...................((((........))))...... ( -21.10) >DroSim_CAF1 65645 90 + 1 GUGAUUGACAACCGUCAAUUGCGGGAAUUCACAUCGCCUAUGGAAUUCCAUUACUCGCCGCUCGUUAUUUGCUACUCGUUGUAAUUGUAC (..((((((....))))))..).(((((((.(((.....))))))))))(((((.((..((.........))....))..)))))..... ( -22.60) >DroEre_CAF1 78436 90 + 1 GUGAUUGACAACCGUCAAUUGCGGGAAUUCACAUCGCCAAUGAAAUUCCAUUACUCGCCGCUCGUUCUUCACUACUCGUUCUAAUUGUAC (..((((((....))))))..)((((.((((.........)))).))))......................................... ( -15.70) >DroYak_CAF1 72911 90 + 1 GUGAUUGACAACCGUCAAUUGCGGGAAUUCACAUCGCCGAUGGAAUUCCAUUACUCGCCGCUCGUUAUUCGCUACUCGUUUUCAUUUUAA (..((((((....))))))..).(((((((.((((...)))))))))))......................................... ( -22.30) >consensus GUGAUUGACAACCGUCAAUUGCGGGAAUUCACAUCGCCUAUGGAAUUCCAUUACUCGCCGCUCGUUAUUUGCUACUCGUUGUAAUUGUAC (..((((((....))))))..).(((((((.(((.....))))))))))......................................... (-18.94 = -19.14 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:09:00 2006