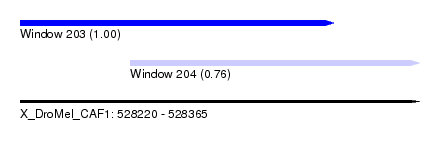
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 528,220 – 528,365 |
| Length | 145 |
| Max. P | 0.997055 |

| Location | 528,220 – 528,334 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.08 |
| Mean single sequence MFE | -21.16 |
| Consensus MFE | -13.90 |
| Energy contribution | -15.58 |
| Covariance contribution | 1.68 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.66 |
| SVM decision value | 2.79 |
| SVM RNA-class probability | 0.997055 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 528220 114 + 22224390 UUUAACUGCUUCCAACGCUUCCUCCCACUGACGCAUAAAUCUGCAGCUGCGUGCGACUUUUUCCACCCACAAAAGUCCACAAAAGCCGUCUUAAGCAUUUAUCAUUUACCAUUU------ ..(((.(((((...(((...............(((......))).(((..(((.(((((((.........))))))))))...))))))...))))).))).............------ ( -20.50) >DroSec_CAF1 14925 113 + 1 UUUAACUGCUUCCAACGCUUCCUCCCACUGACGCAUAAAUCUGCGGCUGCGUGCGACUUUUGCCACCCACUAAAGUCC-CAAAAGCCGUCUUAAGCAUUUAUCAUUUACCAUUU------ ..(((.(((((.....((.((........)).))........((((((...((.((((((...........)))))).-))..))))))...))))).))).............------ ( -22.70) >DroSim_CAF1 15244 113 + 1 UUUAACUGCUUCCAACGCUUCCUCCCACUGACGCAUAAAUCUGCGGCUGCGUGCGACUUUUGCCACCCACAAAAGUCC-CAAAAGCCGUCUAAAGCAUUUAUCAUUUACCAUUU------ ..(((.(((((.....((.((........)).))........((((((...((.((((((((.......)))))))).-))..))))))...))))).))).............------ ( -26.80) >DroEre_CAF1 19949 102 + 1 ------------------CUUCCCCCACUGGCGCAUAAAUCUGCGGCUGCGUGUGACUUUUGCCACCCACAAGCGUCCACAAAAGCCCUCUUCAUCAUGAAUCAUUUACCAUUUACCAUU ------------------..........(((.(((......)))((((..(((.(((.((((.......)))).))))))...)))).....................)))......... ( -18.40) >DroYak_CAF1 14380 89 + 1 ------------------UUUCUACAA------CAUAAAUCUGCGGCUGCGUGCGACU-UUGCCACCCACAAACGUCCACAAAAGCCGUCUUCAUCAUUAAUCAUUUACCACUU------ ------------------.........------.........((((((..(((.((((-(((.......)))).))))))...)))))).........................------ ( -17.40) >consensus UUUAACUGCUUCCAACGCUUCCUCCCACUGACGCAUAAAUCUGCGGCUGCGUGCGACUUUUGCCACCCACAAAAGUCCACAAAAGCCGUCUUAAGCAUUUAUCAUUUACCAUUU______ ..........................................((((((..(((.((((((((.......)))))))))))...))))))............................... (-13.90 = -15.58 + 1.68)


| Location | 528,260 – 528,365 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.90 |
| Mean single sequence MFE | -17.42 |
| Consensus MFE | -13.90 |
| Energy contribution | -15.58 |
| Covariance contribution | 1.68 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.759206 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
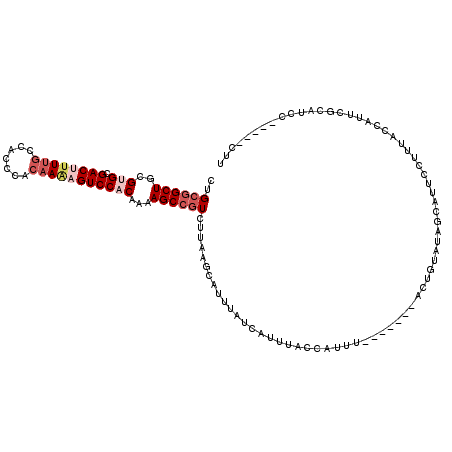
>X_DroMel_CAF1 528260 105 + 22224390 CUGCAGCUGCGUGCGACUUUUUCCACCCACAAAAGUCCACAAAAGCCGUCUUAAGCAUUUAUCAUUUACCAUUU-------ACUGCAUCGCAUUCCUUUACCAUUCGCAUCC-------- .(((((....(((.(((((((.........))))))))))....((........))..................-------.))))).........................-------- ( -15.80) >DroSec_CAF1 14965 112 + 1 CUGCGGCUGCGUGCGACUUUUGCCACCCACUAAAGUCC-CAAAAGCCGUCUUAAGCAUUUAUCAUUUACCAUUU-------ACUGUAUAGCAUUCCUUUACCAUUCGCAUCCCCUUUCUU .(((((((...((.((((((...........)))))).-))..))))...........................-------...(((.((.....)).))).....)))........... ( -16.70) >DroSim_CAF1 15284 112 + 1 CUGCGGCUGCGUGCGACUUUUGCCACCCACAAAAGUCC-CAAAAGCCGUCUAAAGCAUUUAUCAUUUACCAUUU-------ACUGUAUAGCAUUCCUUUACCAUUCGCAUCCCCUUUCUU .(((((((...((.((((((((.......)))))))).-))..))))...........................-------...(((.((.....)).))).....)))........... ( -20.70) >DroEre_CAF1 19971 109 + 1 CUGCGGCUGCGUGUGACUUUUGCCACCCACAAGCGUCCACAAAAGCCCUCUUCAUCAUGAAUCAUUUACCAUUUACCAUUUGCUGUAUACCAUUGCUAUACCAUUC-----------CUC ..((((((..(((.(((.((((.......)))).))))))...))))...(((.....)))....................)).(((((.......))))).....-----------... ( -16.50) >DroYak_CAF1 14396 101 + 1 CUGCGGCUGCGUGCGACU-UUGCCACCCACAAACGUCCACAAAAGCCGUCUUCAUCAUUAAUCAUUUACCACUU-------ACUGUACACCAUUGCCAUACCAUUC-----------CUU ..((((((..(((.((((-(((.......)))).))))))...)))))).........................-------.........................-----------... ( -17.40) >consensus CUGCGGCUGCGUGCGACUUUUGCCACCCACAAAAGUCCACAAAAGCCGUCUUAAGCAUUUAUCAUUUACCAUUU_______ACUGUAUAGCAUUCCUUUACCAUUCGCAUCC_____CUU ..((((((..(((.((((((((.......)))))))))))...))))))....................................................................... (-13.90 = -15.58 + 1.68)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:36:08 2006