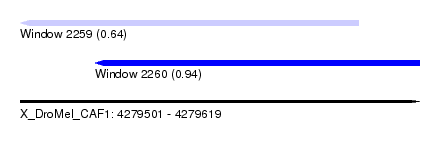
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,279,501 – 4,279,619 |
| Length | 118 |
| Max. P | 0.940199 |

| Location | 4,279,501 – 4,279,601 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 80.38 |
| Mean single sequence MFE | -19.52 |
| Consensus MFE | -11.70 |
| Energy contribution | -11.70 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.636413 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4279501 100 - 22224390 GUUGUUUUUUUUUUUUUUUGUUUGAGGGGAAACUUGGAUUUUUACCGGCGACGGAAACGCUUACUUAAUUAGCCGAACCAAAAAAAAAAAAAAAGAAAAA .((.((((((((((((((((((((.(((....))).........(((....)))....(((.........))))))).)))))))))))))))).))... ( -24.40) >DroSec_CAF1 65875 82 - 1 GUUUU---------------UUUGAGGGGAAACUUGGAUUUAUACCGGCGACGGAAGCGCUUACUUAAUUAGCCGAACCAAAAAA---AAAAAAGAAAAC .((((---------------(((..(((....)((((..(((....((((.(....)))))....)))....)))).))..))))---)))......... ( -17.20) >DroSim_CAF1 60433 82 - 1 GUUUU---------------UUUGAGGGGAAACUUGGAUUUAUACCGGCGACGGAAGCGCUUACUUAAUUAGCCGAACCAAAAAA---AAAAAAGAAAAC .((((---------------(((..(((....)((((..(((....((((.(....)))))....)))....)))).))..))))---)))......... ( -17.20) >DroEre_CAF1 72915 83 - 1 -------UUUUCUU------UUUGAGGGGAAACUUGGAUUUUUACCGGCAACGGAAGCGCUUACUUAAUUAGCCGAACCAAAAAAAA-A---AAAAAUAC -------((((.((------(((..(((....)((((.......(((....)))(((......)))......)))).))..))))).-)---)))..... ( -14.40) >DroYak_CAF1 67458 98 - 1 UUUAUUAUUUUCUUUUUUU-GUUGAGGGGAAACUUGGAUUUUUACCGGCGACGGAAGCGCUUACUUAAUUAGCCGAACCAAACAAGA-AAAAAAUAAUAC ..(((((((((..((((((-(((..(((....)((((....(((..((((.(....)))))....)))....)))).)).)))))))-))))))))))). ( -24.40) >consensus GUUUU__UUUU_UU______UUUGAGGGGAAACUUGGAUUUUUACCGGCGACGGAAGCGCUUACUUAAUUAGCCGAACCAAAAAA_A_AAAAAAGAAAAC .........................(((....)))((.......(((....)))....(((.........)))....))..................... (-11.70 = -11.70 + 0.00)


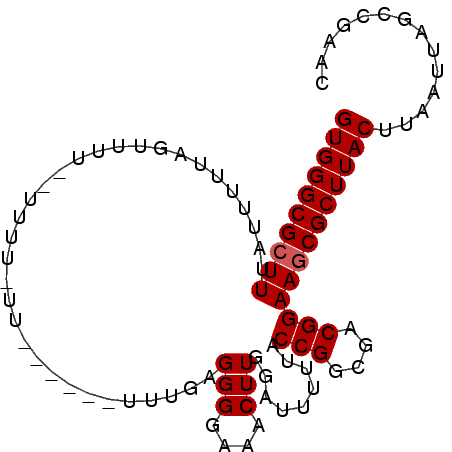
| Location | 4,279,523 – 4,279,619 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 83.44 |
| Mean single sequence MFE | -23.68 |
| Consensus MFE | -22.00 |
| Energy contribution | -22.20 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.940199 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4279523 96 - 22224390 GUGGGCGCUUUAUUUUUAGUUGUUUUUUUUUUUUUUUGUUUGAGGGGAAACUUGGAUUUUUACCGGCGACGGAAACGCUUACUUAAUUAGCCGAAC (((((((..............................(((..((......))..))).....(((....)))...))))))).............. ( -19.20) >DroSec_CAF1 65894 81 - 1 GUGGGCGCUUUAUUUUUAGUUUU---------------UUUGAGGGGAAACUUGGAUUUAUACCGGCGACGGAAGCGCUUACUUAAUUAGCCGAAC ((((((((((...((..((((((---------------((....))))))))..))......(((....))))))))))))).............. ( -24.30) >DroSim_CAF1 60452 81 - 1 GUGGGCGCUUUAUUUUUAGUUUU---------------UUUGAGGGGAAACUUGGAUUUAUACCGGCGACGGAAGCGCUUACUUAAUUAGCCGAAC ((((((((((...((..((((((---------------((....))))))))..))......(((....))))))))))))).............. ( -24.30) >DroEre_CAF1 72933 80 - 1 GUGGGCGCUUUAUUU----------UUUUCUU------UUUGAGGGGAAACUUGGAUUUUUACCGGCAACGGAAGCGCUUACUUAAUUAGCCGAAC ((((((((((...((----------((..(((------...)))..))))............(((....))))))))))))).............. ( -25.20) >DroYak_CAF1 67479 93 - 1 GUGGGCGCUUUAUUU--AUUUAUUAUUUUCUUUUUUU-GUUGAGGGGAAACUUGGAUUUUUACCGGCGACGGAAGCGCUUACUUAAUUAGCCGAAC ((((((((((.....--(((((...(((((((((...-...)))))))))..))))).....(((....))))))))))))).............. ( -25.40) >consensus GUGGGCGCUUUAUUUUUAGUUUU__UUUU_UU______UUUGAGGGGAAACUUGGAUUUUUACCGGCGACGGAAGCGCUUACUUAAUUAGCCGAAC ((((((((((.................................(((....))).........(((....))))))))))))).............. (-22.00 = -22.20 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:08:56 2006