
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,231,520 – 4,231,611 |
| Length | 91 |
| Max. P | 0.769237 |

| Location | 4,231,520 – 4,231,611 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 77.77 |
| Mean single sequence MFE | -29.42 |
| Consensus MFE | -24.68 |
| Energy contribution | -24.48 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.06 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.745464 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4231520 91 + 22224390 -----UAA----AUAUAUCCCCACUGGCAU--GAAGGAUGACGCCAAUUGGCCUGCGUUGCUUAGCGCCAACGAAAGGCGCUGCGGCCAACACACA----------GUCAGC -----...----...........(((((.(--(..((......))..((((((.((...)).(((((((.......))))))).))))))....))----------))))). ( -28.10) >DroPse_CAF1 19769 110 + 1 UACACCGAAAAGAAGUGGGACCCUGGGCAU--GAAGGAUGACGCCAAUUGGCCUGAGUUGCUUAGCGCCAACGCCAGGCGCUUCAGCCAACACACACAGCCCCAUUGCCAGC .............((((((...((((((..--..........)))..(((((.((((.(((((.(((....))).))))).)))))))))......))).))))))...... ( -32.50) >DroEre_CAF1 24484 91 + 1 -----UAA----AUAUAUCCCCACUGGCAU--GAAGGAUGACGCCAAUUGGCCUGCGUUGCUUAGCGCCAACGAAAGGCGCUGCGGCCAACACACA----------GUCAGC -----...----...........(((((.(--(..((......))..((((((.((...)).(((((((.......))))))).))))))....))----------))))). ( -28.10) >DroYak_CAF1 18630 91 + 1 -----UAA----AUAUAUCCACACUGGCAU--GAAGGAUGACGCCAAUUGGCCUGCGUUGCUUAGCGCCAACGAAAGGCGCUGCGGCCAACACACA----------GUCAGC -----...----...........(((((.(--(..((......))..((((((.((...)).(((((((.......))))))).))))))....))----------))))). ( -28.10) >DroAna_CAF1 21744 106 + 1 -----CAAAAAUAUAUAUCCCCACCGGCAUUUUAAGGAUGACGCCAAUUGGCCUGCGUUGCUUAGCGCCAAAGCCAGGCGCUACGGCCAACACACACACACACUUAGUCAG- -----....................(((....((((..((..(((....))).((.(((((((((((((.......))))))).)).)))).))....))..)))))))..- ( -27.20) >DroPer_CAF1 19414 110 + 1 UACACCGAAAAGAAGUGGGACCCUGGGCAU--GAAGGAUGACGCCAAUUGGCCUGAGUUGCUUAGCGCCAACGCCAGGCGCUUCAGCCAACACACACAGCCCCAUUGCCAGC .............((((((...((((((..--..........)))..(((((.((((.(((((.(((....))).))))).)))))))))......))).))))))...... ( -32.50) >consensus _____CAA____AUAUAUCCCCACUGGCAU__GAAGGAUGACGCCAAUUGGCCUGCGUUGCUUAGCGCCAACGAAAGGCGCUGCGGCCAACACACA__________GUCAGC .........................(((..............(((....))).((.(((((((((((((.......))))))).)).)))).))............)))... (-24.68 = -24.48 + -0.19)

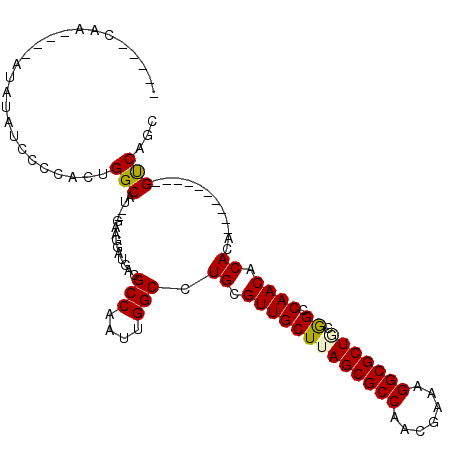

| Location | 4,231,520 – 4,231,611 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 77.77 |
| Mean single sequence MFE | -36.22 |
| Consensus MFE | -25.90 |
| Energy contribution | -25.04 |
| Covariance contribution | -0.86 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.769237 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4231520 91 - 22224390 GCUGAC----------UGUGUGUUGGCCGCAGCGCCUUUCGUUGGCGCUAAGCAACGCAGGCCAAUUGGCGUCAUCCUUC--AUGCCAGUGGGGAUAUAU----UUA----- ...(((----------.((..(((((((..((((((.......))))))..((...)).)))))))..)))))(((((.(--((....))))))))....----...----- ( -33.40) >DroPse_CAF1 19769 110 - 1 GCUGGCAAUGGGGCUGUGUGUGUUGGCUGAAGCGCCUGGCGUUGGCGCUAAGCAACUCAGGCCAAUUGGCGUCAUCCUUC--AUGCCCAGGGUCCCACUUCUUUUCGGUGUA (((((.(((((((((.((...(((((((..((.((.(((((....))))).))..))..))))))).(((((........--))))))).)))))))....)).)))))... ( -40.00) >DroEre_CAF1 24484 91 - 1 GCUGAC----------UGUGUGUUGGCCGCAGCGCCUUUCGUUGGCGCUAAGCAACGCAGGCCAAUUGGCGUCAUCCUUC--AUGCCAGUGGGGAUAUAU----UUA----- ...(((----------.((..(((((((..((((((.......))))))..((...)).)))))))..)))))(((((.(--((....))))))))....----...----- ( -33.40) >DroYak_CAF1 18630 91 - 1 GCUGAC----------UGUGUGUUGGCCGCAGCGCCUUUCGUUGGCGCUAAGCAACGCAGGCCAAUUGGCGUCAUCCUUC--AUGCCAGUGUGGAUAUAU----UUA----- .....(----------(((((....((...((((((.......))))))..)).)))))).(((((((((((........--)))))))).)))......----...----- ( -33.30) >DroAna_CAF1 21744 106 - 1 -CUGACUAAGUGUGUGUGUGUGUUGGCCGUAGCGCCUGGCUUUGGCGCUAAGCAACGCAGGCCAAUUGGCGUCAUCCUUAAAAUGCCGGUGGGGAUAUAUAUUUUUG----- -......((((((((((.(((((((....(((((((.......)))))))..)))))))..((.((((((((..........)))))))).)).))))))))))...----- ( -37.20) >DroPer_CAF1 19414 110 - 1 GCUGGCAAUGGGGCUGUGUGUGUUGGCUGAAGCGCCUGGCGUUGGCGCUAAGCAACUCAGGCCAAUUGGCGUCAUCCUUC--AUGCCCAGGGUCCCACUUCUUUUCGGUGUA (((((.(((((((((.((...(((((((..((.((.(((((....))))).))..))..))))))).(((((........--))))))).)))))))....)).)))))... ( -40.00) >consensus GCUGAC__________UGUGUGUUGGCCGCAGCGCCUGGCGUUGGCGCUAAGCAACGCAGGCCAAUUGGCGUCAUCCUUC__AUGCCAGUGGGGAUAUAU____UUA_____ .................(((((((((((((((((((.......)))))).......)).))))))).(((((..........))))).......)))).............. (-25.90 = -25.04 + -0.86)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:08:45 2006