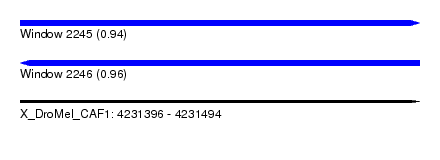
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,231,396 – 4,231,494 |
| Length | 98 |
| Max. P | 0.958399 |

| Location | 4,231,396 – 4,231,494 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 76.93 |
| Mean single sequence MFE | -23.47 |
| Consensus MFE | -15.85 |
| Energy contribution | -16.05 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938248 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4231396 98 + 22224390 UUAUCUUAAAAAUGUGCUACAAUUAUG-AAUUUAUUGCUUUUCUUGCAGUGCCUUAAAGCAACAUAAGGAUGAAGGCGGCAAGUGCCGCCAUACACACA (((((((....(((((((.((....))-....((((((.......))))))......))).)))).))))))).((((((....))))))......... ( -26.30) >DroSec_CAF1 18674 97 + 1 -UGUCUUAGAAAUGUGCUACACUUAUG-AAUUUAUUGCUUUUUUUGCAGUGCCUUAAAGCAACAUGAGGAUGAAGGCGGCAAGUGCCGCCAUACACACA -...........((((.....((((((-.....(((((.......)))))((......))..))))))......((((((....))))))...)))).. ( -28.00) >DroSim_CAF1 24350 97 + 1 -UGUCUUAGAAAUGUGCUACAAUUAUG-AAUUUAUUGCUUUUUUUGCAGUGCCUUAAAGCAACAUGAGGAUGAAGGCGGCAAGUGCCGCCAUACACACA -.(((((....(((((((.((....))-....((((((.......))))))......))).)))).)))))...((((((....))))))......... ( -26.60) >DroEre_CAF1 24361 97 + 1 -UAUUUUUGAAAUGCCCUACUUUUAGG-UAUUAAUUGUUUUUCUUGCAGUGCCCUCCAGCAACAUGAGGACGAAGGCUGCAAGUGCCGCCACACACACA -.........((((((.........))-))))..((((((((..((...(((......))).)).)))))))).(((.((....)).)))......... ( -19.00) >DroYak_CAF1 18506 98 + 1 -UAUUUUAGAAAUUCACUACUAUUAUGUUAUUUAUUGUAUUUUUUGCAGUGCCUUUAAGCAACAUGAGGAUAAAGGCGGCAAGUGCCGCCACACACACA -.....................(((((((....((((((.....))))))((......))))))))).......((((((....))))))......... ( -23.20) >DroAna_CAF1 21630 74 + 1 ---------------------GUAAUU-GAUUU---AUAUUUCGUACAGUGCUUGCCACAAAAUUAAGGAUGAAGGCGGCAAGUGCCGCCACACACACA ---------------------(((...-(((..---..)))...))).(((..((((..........)).....((((((....)))))).))..))). ( -17.70) >consensus _UAUCUUAGAAAUGUGCUACAAUUAUG_AAUUUAUUGCUUUUCUUGCAGUGCCUUAAAGCAACAUGAGGAUGAAGGCGGCAAGUGCCGCCACACACACA ...................................(((.......)))(((((((..........)))).....((((((....))))))...)))... (-15.85 = -16.05 + 0.20)



| Location | 4,231,396 – 4,231,494 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 76.93 |
| Mean single sequence MFE | -24.73 |
| Consensus MFE | -15.06 |
| Energy contribution | -15.15 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.958399 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4231396 98 - 22224390 UGUGUGUAUGGCGGCACUUGCCGCCUUCAUCCUUAUGUUGCUUUAAGGCACUGCAAGAAAAGCAAUAAAUU-CAUAAUUGUAGCACAUUUUUAAGAUAA .((((((..((((((....))))))..........(((((((((...((...))....)))))))))....-..........))))))........... ( -27.90) >DroSec_CAF1 18674 97 - 1 UGUGUGUAUGGCGGCACUUGCCGCCUUCAUCCUCAUGUUGCUUUAAGGCACUGCAAAAAAAGCAAUAAAUU-CAUAAGUGUAGCACAUUUCUAAGACA- .((((((..((((((....))))))..........(((((((((...((...))....)))))))))....-..........))))))..........- ( -27.90) >DroSim_CAF1 24350 97 - 1 UGUGUGUAUGGCGGCACUUGCCGCCUUCAUCCUCAUGUUGCUUUAAGGCACUGCAAAAAAAGCAAUAAAUU-CAUAAUUGUAGCACAUUUCUAAGACA- .((((((..((((((....))))))..........(((((((((...((...))....)))))))))....-..........))))))..........- ( -27.90) >DroEre_CAF1 24361 97 - 1 UGUGUGUGUGGCGGCACUUGCAGCCUUCGUCCUCAUGUUGCUGGAGGGCACUGCAAGAAAAACAAUUAAUA-CCUAAAAGUAGGGCAUUUCAAAAAUA- ..(((...(((..((.(((((((.....((((((.........)))))).)))))))..............-((((....))))))...)))...)))- ( -23.40) >DroYak_CAF1 18506 98 - 1 UGUGUGUGUGGCGGCACUUGCCGCCUUUAUCCUCAUGUUGCUUAAAGGCACUGCAAAAAAUACAAUAAAUAACAUAAUAGUAGUGAAUUUCUAAAAUA- (((((.((..(.(((....)))(((((((..(.......)..))))))).)..))....)))))..................................- ( -21.40) >DroAna_CAF1 21630 74 - 1 UGUGUGUGUGGCGGCACUUGCCGCCUUCAUCCUUAAUUUUGUGGCAAGCACUGUACGAAAUAU---AAAUC-AAUUAC--------------------- .(..((...((((((....))))))..))..)...((((((((.((.....))))))))))..---.....-......--------------------- ( -19.90) >consensus UGUGUGUAUGGCGGCACUUGCCGCCUUCAUCCUCAUGUUGCUUUAAGGCACUGCAAAAAAAACAAUAAAUU_CAUAAUUGUAGCACAUUUCUAAGAUA_ .(..((...((((((....))))))..))..)......((((....))))................................................. (-15.06 = -15.15 + 0.09)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:08:44 2006