
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,231,056 – 4,231,188 |
| Length | 132 |
| Max. P | 0.990686 |

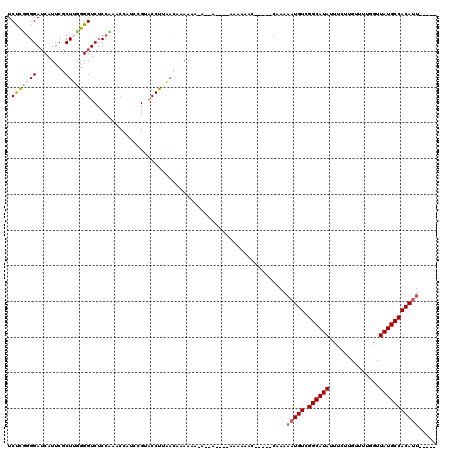
| Location | 4,231,056 – 4,231,162 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.10 |
| Mean single sequence MFE | -20.53 |
| Consensus MFE | -13.36 |
| Energy contribution | -13.42 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.653618 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4231056 106 - 22224390 UCUCGGGGAUCAUUCGCUUGGGGUCUCCUAACCAUCCGUACCUUAACAAAAAAAAAAA----AAAAAAG-----UAAAAAUGUCGGCAUAUUUCUUGUUUUGGUUAUGCCACAUU----- ....(((((((..........)))))))..............................----.......-----....(((((.((((((...((......)).)))))))))))----- ( -19.70) >DroSec_CAF1 18337 107 - 1 UCUCGGGGAUCAUUCGCUUUGGGUCUCCUAACCAUCCGUACCUUAACAAAAAA---AAGAUGAAAAAAA-----CAAAAAUGUCGGCAUAUUUCUGGUUUUGGUUAUGCCACAUU----- ....(((((((..........)))))))....((((.................---..)))).......-----....(((((.((((((...((......)).)))))))))))----- ( -20.41) >DroSim_CAF1 24009 110 - 1 UCUCGGGGAUCAUUCGCUACGGGUCUCCCAACCAUCCGUACCUUAACAAAAAAAAAAAGAAGCAAAAAC-----CAAAAAUGUCGGCAUAUUUCUUGUUUUGGUUAUGCCACAUU----- ....((.((....))(((((((((.........))))))..(((............))).))).....)-----)...(((((.((((((...((......)).)))))))))))----- ( -19.70) >DroEre_CAF1 24055 109 - 1 UCUCUGGGAUCAUUCGCUUGGGGUCUCCAUACCAUCCGAGCCUUGACAAAAAAAA--A----GAAUAAC-----CAAAAAUGUCGGCAUAUUUCUUGUUUCCAUUAUGCCACAUUUCAUU ....(((((....))(((((((((......)))..))))))..............--.----......)-----)).((((((.((((((..............)))))))))))).... ( -23.34) >DroYak_CAF1 18210 97 - 1 UCUCUGGGAUCAUUCGCUUGGGGUCUCCAUACCAUCCGAACCUUAACAAAA---A--A----GGAAAAC-----CAAAUAUGUCGGCAUAUUUCU---------UAUGCCACAUUUCAUU ....(((.....((((..(((..........)))..))))((((.......---)--)----))....)-----))...((((.((((((.....---------))))))))))...... ( -22.20) >DroAna_CAF1 21303 108 - 1 UCCUUUGGAACCUACGCUUGAGGUCUCC---------GUACCUCACAAUAAAA-A--ACGUAAAAAAAAUAUGAAAAAACUGUCGGCAUAUUUCUUGUUUUCAUUAUGCCACAUUUCAUU (((...))).........((((((....---------..))))))........-.--.............((((((....(((.((((((..............))))))))))))))). ( -17.84) >consensus UCUCGGGGAUCAUUCGCUUGGGGUCUCCAAACCAUCCGUACCUUAACAAAAAA_A__A____AAAAAAC_____CAAAAAUGUCGGCAUAUUUCUUGUUUUGGUUAUGCCACAUU_____ .(((((((........))))))).......................................................(((((.((((((..............)))))))))))..... (-13.36 = -13.42 + 0.06)


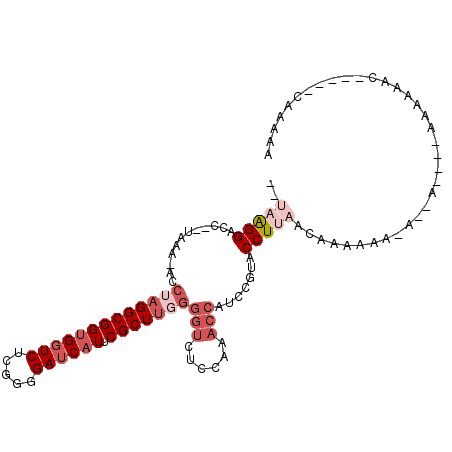
| Location | 4,231,091 – 4,231,188 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 73.59 |
| Mean single sequence MFE | -24.30 |
| Consensus MFE | -15.23 |
| Energy contribution | -17.15 |
| Covariance contribution | 1.92 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.63 |
| SVM decision value | 2.23 |
| SVM RNA-class probability | 0.990686 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4231091 97 - 22224390 --UUAGGUCC--UACAAACCUAGGCGGUGGUCUCGGGGAUCAUUCGCUUGGGGUCUCCUAACCAUCCGUACCUUAACAAAAAAAAAAA----AAAAAAG-----UAAAAA --((((((..--.....))))))((((((((...(((((((..........)))))))..)))).))))...................----.......-----...... ( -23.00) >DroSec_CAF1 18372 97 - 1 --UAAGGACC--UACA-ACCUAGGCGGUGGUCUCGGGGAUCAUUCGCUUUGGGUCUCCUAACCAUCCGUACCUUAACAAAAAA---AAGAUGAAAAAAA-----CAAAAA --(((((.((--((..-...))))(((((((...(((((((..........)))))))..)))).)))..)))))........---.............-----...... ( -23.70) >DroSim_CAF1 24044 100 - 1 --UAAGGACC--UACA-ACCUAGGCGGUGGUCUCGGGGAUCAUUCGCUACGGGUCUCCCAACCAUCCGUACCUUAACAAAAAAAAAAAGAAGCAAAAAC-----CAAAAA --(((((.((--((..-...))))(((((((...(((((((..........)))))))..)))).)))..)))))........................-----...... ( -23.70) >DroEre_CAF1 24095 94 - 1 --AAGGGUCC--UAAA-AACUAGGCGGUGGUCUCUGGGAUCAUUCGCUUGGGGUCUCCAUACCAUCCGAGCCUUGACAAAAAAAA--A----GAAUAAC-----CAAAAA --((((.((.--....-..(((((((((((((.....))))).))))))))(((......)))....)).))))...........--.----.......-----...... ( -24.30) >DroYak_CAF1 18241 91 - 1 --UAGGGCCC--UAAA-AACUAGGCGGUGGUCUCUGGGAUCAUUCGCUUGGGGUCUCCAUACCAUCCGAACCUUAACAAAA---A--A----GGAAAAC-----CAAAUA --..((((((--((..-.....((((((((((.....))))).)))))))))))))..............((((.......---)--)----)).....-----...... ( -23.21) >DroAna_CAF1 21343 97 - 1 AAAAGGGGCGGGGAAA-ACCUAGGCGGUGGUCCUUUGGAACCUACGCUUGAGGUCUCC---------GUACCUCACAAUAAAA-A--ACGUAAAAAAAAUAUGAAAAAAC ...(((..((((((..-.((((((((..(((((...)).)))..))))).))))))))---------)..)))..........-.--.((((.......))))....... ( -27.90) >consensus __UAAGGACC__UAAA_ACCUAGGCGGUGGUCUCGGGGAUCAUUCGCUUGGGGUCUCCAAACCAUCCGUACCUUAACAAAAAA_A__A____AAAAAAC_____CAAAAA ..(((((............(((((((((((((.....)))))).)))))))(((......))).......)))))................................... (-15.23 = -17.15 + 1.92)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:08:42 2006