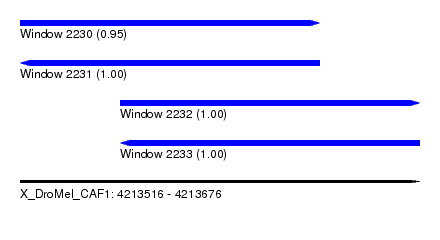
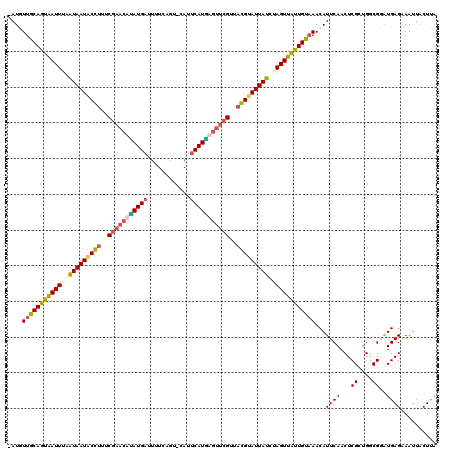
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,213,516 – 4,213,676 |
| Length | 160 |
| Max. P | 0.999999 |

| Location | 4,213,516 – 4,213,636 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.15 |
| Mean single sequence MFE | -23.98 |
| Consensus MFE | -17.76 |
| Energy contribution | -19.20 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.951324 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4213516 120 + 22224390 UAAGUAAUUUCUCAUACGCCAGCGAGUUGAAUGUUUACAACAACUAGAUAAUGCGUAACUACCUAAUGAAUGAACUGAAAAUCAUUAGUUCGAAACGUAUUAUUUAAUUAUUGCAAUAUA ...(((........)))....((((((((........))))...((((((((((((..(........)..((((((((......))))))))..))))))))))))....))))...... ( -23.50) >DroSec_CAF1 1664 118 + 1 CAAGUAAUUCCUCAUCCGCCAGCGAGUUGAAUGUUUACAAUAACUAGAUAAUACCUAACGAACUCAUGAAUG-ACUGAAAAUCAUAUGUUCGAAAGGUAUUACUAAAUUACUGCAACAU- ..(((((((..(((.(((....)).).)))..............(((.((((((((..(((((..((((.(.-......).))))..)))))..)))))))))))))))))).......- ( -25.30) >DroSim_CAF1 1664 118 + 1 CAAGUAAUUCCUCAUCCGCCAGCGAGUUGAAUGUUUACAAUAACUAGAUAAUACCUGACGAACUCAUGAAUG-ACUGAAAAUCAUGUGUUCGAAAGGUAUUACUAAAUUACUGCAAGAU- ..(((((((..(((.(((....)).).)))..............(((.((((((((..(((((.(((((.(.-......).))))).)))))..)))))))))))))))))).......- ( -29.90) >DroEre_CAF1 1696 119 + 1 UGAGCAAUUUCUCACACGCCAACGAGUUGAAUGUUAACAAUAACUUAAUAAUCCGUUUCGCAUUUAUGAAUG-AUUAAAGCACAUAACUUCGAAUCGAAUUAUUAAAUCACUGUAACAUA ((((......)))).....(((....))).((((((.((.....((((((((.((.((((...(((((....-.........)))))...)))).)).)))))))).....)))))))). ( -17.22) >consensus CAAGUAAUUCCUCAUACGCCAGCGAGUUGAAUGUUUACAAUAACUAGAUAAUACCUAACGAACUCAUGAAUG_ACUGAAAAUCAUAAGUUCGAAACGUAUUACUAAAUUACUGCAACAU_ .....................(((.((((........))))...((((((((((((..(((((((((((.(........).)))))))))))..)))))))))))).....)))...... (-17.76 = -19.20 + 1.44)

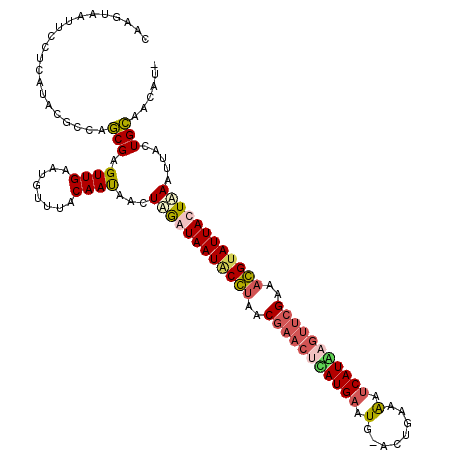

| Location | 4,213,516 – 4,213,636 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.15 |
| Mean single sequence MFE | -34.38 |
| Consensus MFE | -29.75 |
| Energy contribution | -30.25 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.29 |
| Mean z-score | -3.98 |
| Structure conservation index | 0.87 |
| SVM decision value | 3.81 |
| SVM RNA-class probability | 0.999634 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4213516 120 - 22224390 UAUAUUGCAAUAAUUAAAUAAUACGUUUCGAACUAAUGAUUUUCAGUUCAUUCAUUAGGUAGUUACGCAUUAUCUAGUUGUUGUAAACAUUCAACUCGCUGGCGUAUGAGAAAUUACUUA ....((((((((((((.(((((.(((..(...(((((((............)))))))...)..))).))))).))))))))))))...((((...((....))..)))).......... ( -27.60) >DroSec_CAF1 1664 118 - 1 -AUGUUGCAGUAAUUUAGUAAUACCUUUCGAACAUAUGAUUUUCAGU-CAUUCAUGAGUUCGUUAGGUAUUAUCUAGUUAUUGUAAACAUUCAACUCGCUGGCGGAUGAGGAAUUACUUG -...(((((((((((..(((((((((..(((((.(((((........-...))))).)))))..)))))))))..))))))))))).(((((..(.....)..)))))............ ( -36.80) >DroSim_CAF1 1664 118 - 1 -AUCUUGCAGUAAUUUAGUAAUACCUUUCGAACACAUGAUUUUCAGU-CAUUCAUGAGUUCGUCAGGUAUUAUCUAGUUAUUGUAAACAUUCAACUCGCUGGCGGAUGAGGAAUUACUUG -...(((((((((((..(((((((((..(((((.(((((........-...))))).)))))..)))))))))..))))))))))).(((((..(.....)..)))))............ ( -37.70) >DroEre_CAF1 1696 119 - 1 UAUGUUACAGUGAUUUAAUAAUUCGAUUCGAAGUUAUGUGCUUUAAU-CAUUCAUAAAUGCGAAACGGAUUAUUAAGUUAUUGUUAACAUUCAACUCGUUGGCGUGUGAGAAAUUGCUCA .((((((((((((((((((((((((.((((...(((((.........-....)))))...)))).)))))))))))))))))).))))))((((....)))).(.(..(....)..).). ( -35.42) >consensus _AUGUUGCAGUAAUUUAAUAAUACCUUUCGAACAUAUGAUUUUCAGU_CAUUCAUGAGUUCGUUACGUAUUAUCUAGUUAUUGUAAACAUUCAACUCGCUGGCGGAUGAGAAAUUACUUA ....((((((((((((.(((((((((..(((((((((((............)))))))))))..))))))))).))))))))))))...((((...((....))..)))).......... (-29.75 = -30.25 + 0.50)


| Location | 4,213,556 – 4,213,676 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.60 |
| Mean single sequence MFE | -22.00 |
| Consensus MFE | -17.46 |
| Energy contribution | -17.69 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.55 |
| Structure conservation index | 0.79 |
| SVM decision value | 6.69 |
| SVM RNA-class probability | 0.999999 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4213556 120 + 22224390 CAACUAGAUAAUGCGUAACUACCUAAUGAAUGAACUGAAAAUCAUUAGUUCGAAACGUAUUAUUUAAUUAUUGCAAUAUAAUAGCAUAUGCAUAUAAUAUGAAUCUACACACACAGUUUA ....((((((((((((..(........)..((((((((......))))))))..))))))))))))(((((((((.(((......))))))).)))))...................... ( -21.00) >DroSec_CAF1 1704 91 + 1 UAACUAGAUAAUACCUAACGAACUCAUGAAUG-ACUGAAAAUCAUAUGUUCGAAAGGUAUUACUAAAUUACUGCAACAU---AUCAUAUGCAAAG------------------------- ....(((.((((((((..(((((..((((.(.-......).))))..)))))..)))))))))))......((((....---......))))...------------------------- ( -20.20) >DroSim_CAF1 1704 91 + 1 UAACUAGAUAAUACCUGACGAACUCAUGAAUG-ACUGAAAAUCAUGUGUUCGAAAGGUAUUACUAAAUUACUGCAAGAU---AUCAUAUGCAAAG------------------------- ....(((.((((((((..(((((.(((((.(.-......).))))).)))))..)))))))))))......((((....---......))))...------------------------- ( -24.80) >consensus UAACUAGAUAAUACCUAACGAACUCAUGAAUG_ACUGAAAAUCAUAUGUUCGAAAGGUAUUACUAAAUUACUGCAACAU___AUCAUAUGCAAAG_________________________ ....(((.((((((((..(((((.(((((.(........).))))).)))))..)))))))))))......((((.............))))............................ (-17.46 = -17.69 + 0.23)

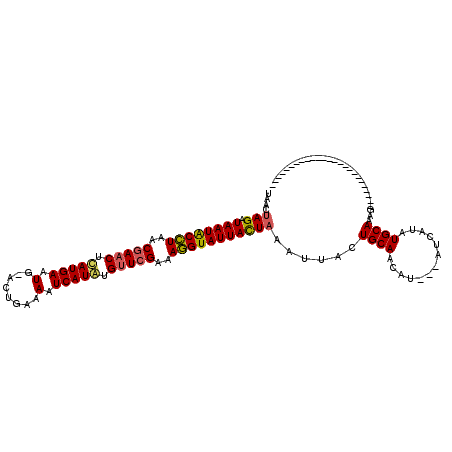
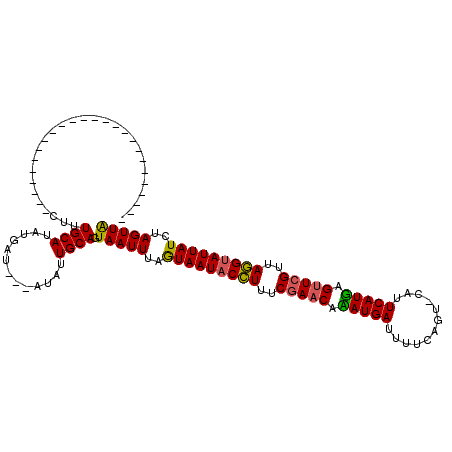
| Location | 4,213,556 – 4,213,676 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.60 |
| Mean single sequence MFE | -26.00 |
| Consensus MFE | -22.29 |
| Energy contribution | -21.85 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.18 |
| Mean z-score | -3.69 |
| Structure conservation index | 0.86 |
| SVM decision value | 6.27 |
| SVM RNA-class probability | 0.999998 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4213556 120 - 22224390 UAAACUGUGUGUGUAGAUUCAUAUUAUAUGCAUAUGCUAUUAUAUUGCAAUAAUUAAAUAAUACGUUUCGAACUAAUGAUUUUCAGUUCAUUCAUUAGGUAGUUACGCAUUAUCUAGUUG .....(((((((((((.......)))))))))))(((.........))).((((((.(((((.(((..(...(((((((............)))))))...)..))).))))).)))))) ( -23.60) >DroSec_CAF1 1704 91 - 1 -------------------------CUUUGCAUAUGAU---AUGUUGCAGUAAUUUAGUAAUACCUUUCGAACAUAUGAUUUUCAGU-CAUUCAUGAGUUCGUUAGGUAUUAUCUAGUUA -------------------------..(((((......---....)))))(((((..(((((((((..(((((.(((((........-...))))).)))))..)))))))))..))))) ( -26.70) >DroSim_CAF1 1704 91 - 1 -------------------------CUUUGCAUAUGAU---AUCUUGCAGUAAUUUAGUAAUACCUUUCGAACACAUGAUUUUCAGU-CAUUCAUGAGUUCGUCAGGUAUUAUCUAGUUA -------------------------..(((((......---....)))))(((((..(((((((((..(((((.(((((........-...))))).)))))..)))))))))..))))) ( -27.70) >consensus _________________________CUUUGCAUAUGAU___AUAUUGCAGUAAUUUAGUAAUACCUUUCGAACAAAUGAUUUUCAGU_CAUUCAUGAGUUCGUUAGGUAUUAUCUAGUUA ............................((((.............)))).(((((..(((((((((..(((((.(((((............))))).)))))..)))))))))..))))) (-22.29 = -21.85 + -0.44)


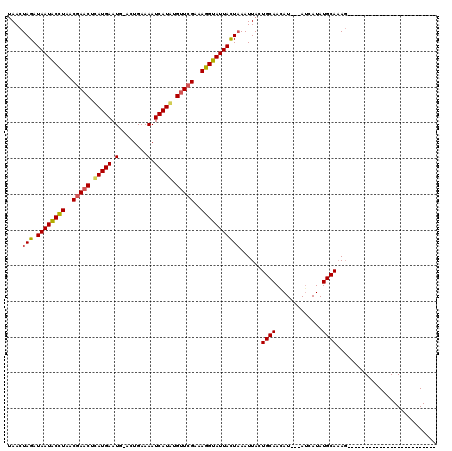
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:08:28 2006