
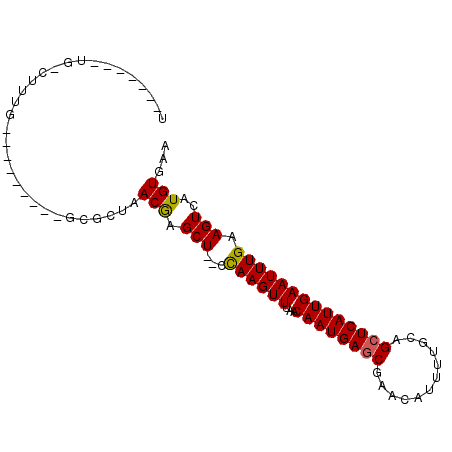
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,192,284 – 4,192,377 |
| Length | 93 |
| Max. P | 0.842756 |

| Location | 4,192,284 – 4,192,377 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 77.35 |
| Mean single sequence MFE | -20.98 |
| Consensus MFE | -13.18 |
| Energy contribution | -12.98 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.716966 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4192284 93 + 22224390 UGUGU--UUGUAUUUGUGUGUCGUCUGGCUAACAAGCUCUCCAAGUUUAACAAUGAGCGAA-AUUUCGCAGCUCAUUGAAUUUGAAGUCAUGUGUA .....--........(..((.(....((((....))))...((((((...((((((((...-........))))))))))))))..).))..)... ( -18.20) >DroVir_CAF1 43671 85 + 1 UGGCUGGCUGGCUUUG---------GCGCUAACGAGCU--UCAAGUUUAACAAUGAGCGAACAUUUUGCAGCUCAUUGAAUUUGAAGUCAUGUGAA ((((..(((......)---------))))))(((.(((--(((((((...((((((((............))))))))))))))))))..)))... ( -25.00) >DroGri_CAF1 32725 80 + 1 U-----ACUGACAUCG---------GCUCUAACGAGCU--UCAAGUUUAACAAUGAGCGAACAUUUUGCAGCUCAUUGAAUUUGAAGUCAUGUGAA (-----((((((....---------((((....))))(--(((((((...((((((((............)))))))))))))))))))).))).. ( -28.20) >DroEre_CAF1 26106 79 + 1 -------------UUGUGU---GUCUGGCUAACAAGCUCUCCAAGUUUAACAAUGAGCGAA-AUUUCGCAGCUCAUUGAAUUUGAAGUCAUGUGUA -------------..(..(---(.((((((....))))...((((((...((((((((...-........)))))))))))))).)).))..)... ( -18.90) >DroWil_CAF1 31922 83 + 1 ------------UGUGUGUGUUGACUGGCUAACGAGCU-UCUAAGUUUAACAAUGAACGAAAAUUUUGCAGCUCAUUGAAUUUGAAGUCAUGUGAA ------------.........(((((((((....))))-...........((((((.(............).)))))).......)))))...... ( -12.00) >DroMoj_CAF1 42759 83 + 1 UGGCU--UUGGCUUUG---------GCGCUAACGAGCU--UCAAGUUUAACAAUGAGCGAACAUUUUGCAGCUCAUUGAAUUUGAAGUCAUGUGAA .....--(((((....---------..))))).((.((--(((((((...((((((((............)))))))))))))))))))....... ( -23.60) >consensus U_______UG_CUUUG_________GCGCUAACGAGCU__CCAAGUUUAACAAUGAGCGAACAUUUUGCAGCUCAUUGAAUUUGAAGUCAUGUGAA ...............................(((.(((...((((((...((((((((............)))))))))))))).)))..)))... (-13.18 = -12.98 + -0.19)

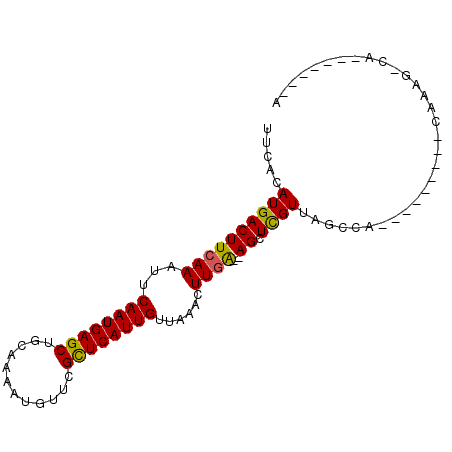
| Location | 4,192,284 – 4,192,377 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 77.35 |
| Mean single sequence MFE | -18.03 |
| Consensus MFE | -10.16 |
| Energy contribution | -9.72 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.842756 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4192284 93 - 22224390 UACACAUGACUUCAAAUUCAAUGAGCUGCGAAAU-UUCGCUCAUUGUUAAACUUGGAGAGCUUGUUAGCCAGACGACACACAAAUACAA--ACACA .........((((((...((((((((........-...))))))))......)))))).(((....)))....................--..... ( -16.20) >DroVir_CAF1 43671 85 - 1 UUCACAUGACUUCAAAUUCAAUGAGCUGCAAAAUGUUCGCUCAUUGUUAAACUUGA--AGCUCGUUAGCGC---------CAAAGCCAGCCAGCCA ..........(((((...((((((((.((.....))..))))))))......))))--)(((.(((.((..---------....)).))).))).. ( -20.70) >DroGri_CAF1 32725 80 - 1 UUCACAUGACUUCAAAUUCAAUGAGCUGCAAAAUGUUCGCUCAUUGUUAAACUUGA--AGCUCGUUAGAGC---------CGAUGUCAGU-----A ...((.((((.((.....((((((((.((.....))..))))))))..........--.((((....))))---------.)).))))))-----. ( -22.90) >DroEre_CAF1 26106 79 - 1 UACACAUGACUUCAAAUUCAAUGAGCUGCGAAAU-UUCGCUCAUUGUUAAACUUGGAGAGCUUGUUAGCCAGAC---ACACAA------------- .........((((((...((((((((........-...))))))))......)))))).(((....))).....---......------------- ( -16.20) >DroWil_CAF1 31922 83 - 1 UUCACAUGACUUCAAAUUCAAUGAGCUGCAAAAUUUUCGUUCAUUGUUAAACUUAGA-AGCUCGUUAGCCAGUCAACACACACA------------ ......(((((.......((((((((............))))))))...........-.(((....))).))))).........------------ ( -12.70) >DroMoj_CAF1 42759 83 - 1 UUCACAUGACUUCAAAUUCAAUGAGCUGCAAAAUGUUCGCUCAUUGUUAAACUUGA--AGCUCGUUAGCGC---------CAAAGCCAA--AGCCA .....((((((((((...((((((((.((.....))..))))))))......))))--)).))))..((..---------....))...--..... ( -19.50) >consensus UUCACAUGACUUCAAAUUCAAUGAGCUGCAAAAUGUUCGCUCAUUGUUAAACUUGA__AGCUCGUUAGCCA_________CAAAG_CA_______A .....((((((((((...((((((((............))))))))......))))..)).))))............................... (-10.16 = -9.72 + -0.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:08:19 2006