
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,187,913 – 4,188,091 |
| Length | 178 |
| Max. P | 0.924069 |

| Location | 4,187,913 – 4,188,029 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 89.43 |
| Mean single sequence MFE | -39.47 |
| Consensus MFE | -31.42 |
| Energy contribution | -32.05 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.924069 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4187913 116 - 22224390 CGU--GAGAGUAUGAAAAUAUGCAAAGGGCAGAACACAAUUUGCGUGUUGUGAUUCGACAAAGGUUUUGUGAAAAACCUUUGGCCUCACCGUGGUUUUCCGGCAUGGGAAAGCGGCCC .((--(((.((((....)))).)....(((.(((((((((......)))))).)))..((((((((((....))))))))))))))))).((.(((((((......))))))).)).. ( -41.40) >DroSec_CAF1 19564 116 - 1 CGU--GAGGGUAUGAAAAUAUGCAAAGGGCAGAACACAAUUUGCGUGUUGUGAUUCGACAAAGGUUUUGCGAAAAACCUUUGGCCUCACAGUGCUUUUCCGGCAUGGGAAAGCGGCCC .((--(((.((((....)))).)....(((.(((((((((......)))))).)))..((((((((((....)))))))))))))))))..(((((((((.....))))))))).... ( -43.80) >DroEre_CAF1 21067 114 - 1 CGU--GAGGGUAUGAAAAUAUGCAAAGGGGAAAACACAAUUUGCGUGUUGUGAUUCGACAAAGGUU-UGCGAG-AACCUUUGGCCUCGUGGUGGUUUCUCGGCAUGGAAAAGCGGCCC ...--((((...((((((((((((((..(.......)..))))))))))....)))).((((((((-(....)-)))))))).))))..(((.((((.((......)).)))).))). ( -36.40) >DroYak_CAF1 24145 116 - 1 CGUAUGAGGGCAUGAAAAUAUGCAAAGGGCAGCACACAAUUUGCGUGUUGUGAUUCGACGAAGGUU-UGCGAG-AACCUUUGGCCUCGUGGUGGUUUCUCGACAUGGUAAAGUGGCCC .......((((.........(((.....)))...(((..(((((((((((.((..(..((((((((-(....)-))))))))(((....))).)..)).)))))).)))))))))))) ( -36.30) >consensus CGU__GAGGGUAUGAAAAUAUGCAAAGGGCAGAACACAAUUUGCGUGUUGUGAUUCGACAAAGGUU_UGCGAA_AACCUUUGGCCUCACGGUGGUUUCCCGGCAUGGGAAAGCGGCCC .....(((.((.((((((((((((((..(.......)..))))))))))....)))).((((((((((....)))))))))))))))..(((.(((((((......))))))).))). (-31.42 = -32.05 + 0.63)


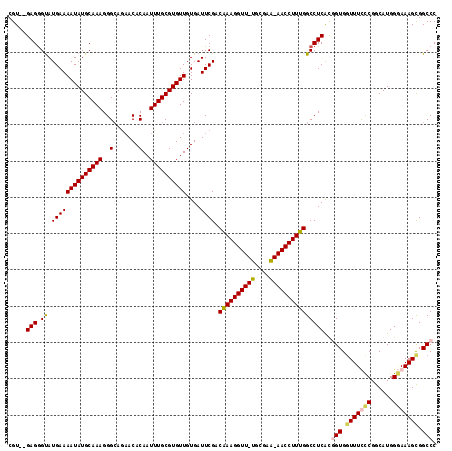
| Location | 4,187,953 – 4,188,065 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 92.90 |
| Mean single sequence MFE | -28.60 |
| Consensus MFE | -20.83 |
| Energy contribution | -22.45 |
| Covariance contribution | 1.62 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.539193 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4187953 112 + 22224390 GGUUUUUCACAAAACCUUUGUCGAAUCACAACACGCAAAUUGUGUUCUGCCCUUUGCAUAUUUUCAUACUCUC--ACGGAGCGUGGAAACCACUGAAAAGUGGAUGUACUAAUU ((((((....))))))...((.(((.(((((........)))))))).))....(((((.((((((..(((..--...)))..))))))(((((....))))))))))...... ( -29.30) >DroSec_CAF1 19604 112 + 1 GGUUUUUCGCAAAACCUUUGUCGAAUCACAACACGCAAAUUGUGUUCUGCCCUUUGCAUAUUUUCAUACCCUC--ACGGAGCGUGAAAACCACUGAAAAGUGGAUGUACUAAUU ((((((....))))))...((.(((.(((((........)))))))).))....(((((.((((((..(((..--..)).)..))))))(((((....))))))))))...... ( -27.70) >DroEre_CAF1 21107 110 + 1 GGUU-CUCGCA-AACCUUUGUCGAAUCACAACACGCAAAUUGUGUUUUCCCCUUUGCAUAUUUUCAUACCCUC--ACGGAGCGUGAAAACCACUGAAAAGUGGAUGUACUAAUU ((((-(..(((-(....)))).)))))..((((((.....))))))........(((((.((((((..(((..--..)).)..))))))(((((....))))))))))...... ( -26.80) >DroYak_CAF1 24185 112 + 1 GGUU-CUCGCA-AACCUUCGUCGAAUCACAACACGCAAAUUGUGUGCUGCCCUUUGCAUAUUUUCAUGCCCUCAUACGGAGCGUGAAAGCCACUGAAAAGUGUAUGUACUAAUU ((((-(..((.-.......)).))))).((.(((((.....))))).)).....(((((((((((((((((......)).))))))))).((((....))))))))))...... ( -30.60) >consensus GGUU_CUCGCA_AACCUUUGUCGAAUCACAACACGCAAAUUGUGUUCUGCCCUUUGCAUAUUUUCAUACCCUC__ACGGAGCGUGAAAACCACUGAAAAGUGGAUGUACUAAUU (((((......)))))...((.(((.(((((........)))))))).))....(((((.(((((((.(((......)).).)))))))(((((....))))))))))...... (-20.83 = -22.45 + 1.62)

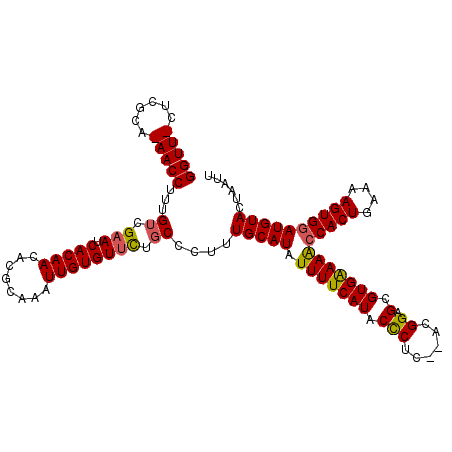
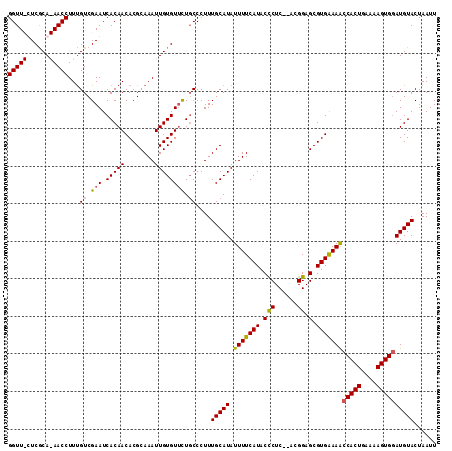
| Location | 4,187,993 – 4,188,091 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 85.69 |
| Mean single sequence MFE | -26.32 |
| Consensus MFE | -18.34 |
| Energy contribution | -19.52 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.751472 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4187993 98 - 22224390 -----AUCGCU---------CACGAUGACACCGAUAAUUGAAUUAGUACAUCCACUUUUCAGUGGUUUCCACGCUCCGU--GAGAGUAUGAAAAUAUGCAAAGGGCAGAACACA -----...(((---------(..((((..((..((......))..)).))))...((((((((((...))))(((.(..--..)))).))))))........))))........ ( -19.80) >DroSec_CAF1 19644 103 - 1 UUUAAAUCGCU---------CACGAUGACACUGAUAAUUGAAUUAGUACAUCCACUUUUCAGUGGUUUUCACGCUCCGU--GAGGGUAUGAAAAUAUGCAAAGGGCAGAACACA ........(((---------(..((((..((((((......)))))).))))((((....))))(((((((..(.((..--..)))..))))))).......))))........ ( -27.80) >DroEre_CAF1 21145 98 - 1 -----GUCGCC---------CAUGAUGAUACUGAUAAUUGAAUUAGUACAUCCACUUUUCAGUGGUUUUCACGCUCCGU--GAGGGUAUGAAAAUAUGCAAAGGGGAAAACACA -----((..((---------(..((((.(((((((......)))))))))))..((((.((...(((((((..(.((..--..)))..))))))).)).)))))))...))... ( -31.70) >DroYak_CAF1 24223 109 - 1 -----AUCGCUGAUACAGAUCACGAUGAUACUGAUAAUUCAAUUAGUACAUACACUUUUCAGUGGCUUUCACGCUCCGUAUGAGGGCAUGAAAAUAUGCAAAGGGCAGCACACA -----...((((...(........(((.(((((((......))))))))))....((((((...(((((((..(...)..))))))).))))))........)..))))..... ( -26.00) >consensus _____AUCGCU_________CACGAUGACACUGAUAAUUGAAUUAGUACAUCCACUUUUCAGUGGUUUUCACGCUCCGU__GAGGGUAUGAAAAUAUGCAAAGGGCAGAACACA ........(((............((((.(((((((......)))))))))))..((((.((...(((((((.((((.......)))).))))))).)).)))))))........ (-18.34 = -19.52 + 1.19)

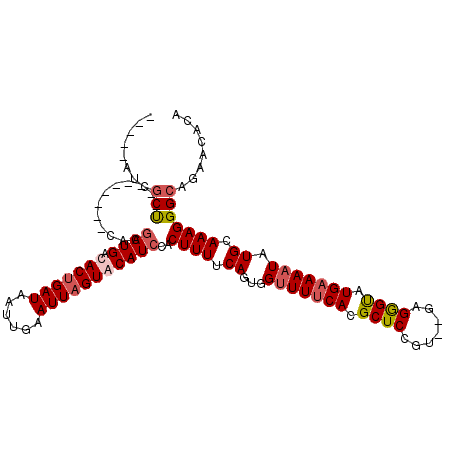
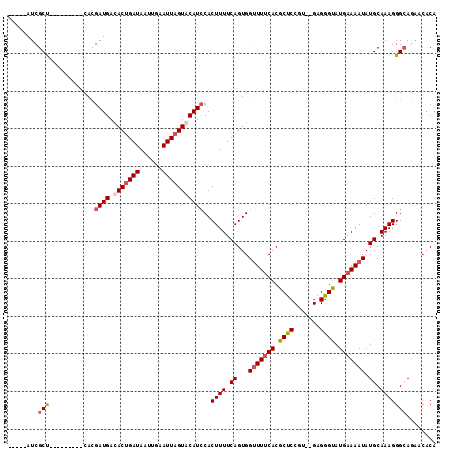
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:08:17 2006