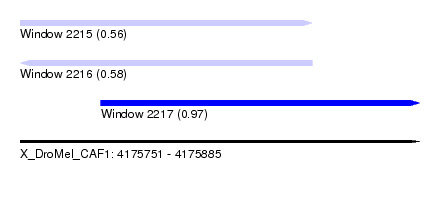
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,175,751 – 4,175,885 |
| Length | 134 |
| Max. P | 0.969065 |

| Location | 4,175,751 – 4,175,849 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 92.86 |
| Mean single sequence MFE | -25.28 |
| Consensus MFE | -17.94 |
| Energy contribution | -18.62 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.558261 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
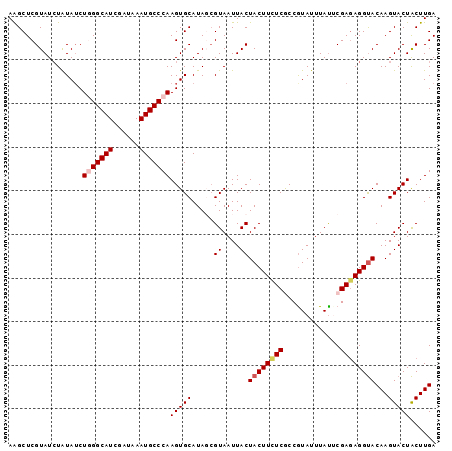
>X_DroMel_CAF1 4175751 98 + 22224390 AGGCUCGUAUCUGUAUCUGGGCAUCGAUAAAUGCCCAAGUGCAUAGAGUAAUUACUACUUUUCGCCAUAUUUAUUCGAGAGGUACAAGUACUACUUGA .(((.......((((..(((((((......)))))))..)))).((((((.....))))))..)))........(((((.((((....)))).))))) ( -30.70) >DroSec_CAF1 8464 98 + 1 AAGCUCGUAUCUAUAUCUGGGCAUCGAUAAAUGCCCAAGUGCAUAGCGUAAUUACUACUUCUCGCCGUAUUUAUACGAGAGGUACAAGUACUACUUGA ..(((.(((.((.....(((((((......))))))))))))..)))........(((((((((...........)))))))))((((.....)))). ( -26.60) >DroSim_CAF1 8864 98 + 1 AAGCUCGUAUAUAUAUCUGGGCAUCGAUAAAUGCCCAAGUGCAUAGCGUAAUUACUACUUCUCGCCGUAUUUAUACGAGAGGUACAAGUACUACUUGA .....(((.(((.((..(((((((......)))))))..)).)))))).......(((((((((...........)))))))))((((.....)))). ( -27.10) >DroEre_CAF1 8451 98 + 1 AAGCUCGUAUCUAUAACUAGGCAUCGAUAAAUGCCCAAGUGCAUAUCGUAAUUACUACUUCUCACCGUAUUUACUCGAUAGGCACAAGUACUGCUUGA ((((..((((.........(((((......)))))...((((.((((((((.(((...........))).))))..)))).))))..)))).)))).. ( -22.80) >DroYak_CAF1 9347 98 + 1 AGGCUCGUAUCUAUAUCUAGGCAUCGAUAAAUGCCCAAGUGCAUAUCGUAAUUACUACUUCUCACCGUAUUUAUUCGAGAGGUACAAGUACUACUUGA .(((...((((.((........)).))))...)))((((((......(((.....((((((((.............))))))))....))))))))). ( -19.22) >consensus AAGCUCGUAUCUAUAUCUGGGCAUCGAUAAAUGCCCAAGUGCAUAGCGUAAUUACUACUUCUCGCCGUAUUUAUUCGAGAGGUACAAGUACUACUUGA .................(((((((......)))))))(((((.....((....))((((((((.............))))))))...)))))...... (-17.94 = -18.62 + 0.68)


| Location | 4,175,751 – 4,175,849 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 92.86 |
| Mean single sequence MFE | -24.52 |
| Consensus MFE | -19.14 |
| Energy contribution | -19.10 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.583679 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
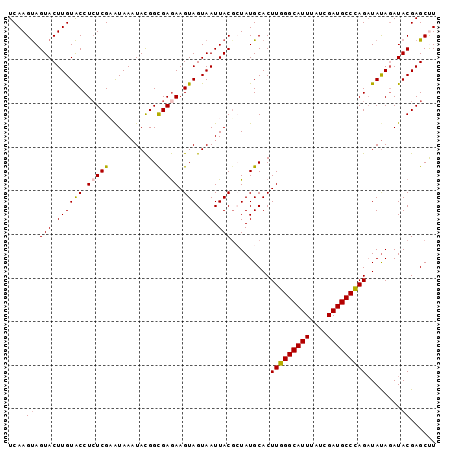
>X_DroMel_CAF1 4175751 98 - 22224390 UCAAGUAGUACUUGUACCUCUCGAAUAAAUAUGGCGAAAAGUAGUAAUUACUCUAUGCACUUGGGCAUUUAUCGAUGCCCAGAUACAGAUACGAGCCU ((.((..(((....)))..)).))........(((.....((((........))))....(((((((((....)))))))))............))). ( -21.80) >DroSec_CAF1 8464 98 - 1 UCAAGUAGUACUUGUACCUCUCGUAUAAAUACGGCGAGAAGUAGUAAUUACGCUAUGCACUUGGGCAUUUAUCGAUGCCCAGAUAUAGAUACGAGCUU ....((((((.((((((.((((((.........)))))).))).))).))).(((((...(((((((((....))))))))).))))).)))...... ( -27.40) >DroSim_CAF1 8864 98 - 1 UCAAGUAGUACUUGUACCUCUCGUAUAAAUACGGCGAGAAGUAGUAAUUACGCUAUGCACUUGGGCAUUUAUCGAUGCCCAGAUAUAUAUACGAGCUU .((((.....)))).....((((((((.(((.((((.((........)).))))......(((((((((....))))))))).))).))))))))... ( -27.20) >DroEre_CAF1 8451 98 - 1 UCAAGCAGUACUUGUGCCUAUCGAGUAAAUACGGUGAGAAGUAGUAAUUACGAUAUGCACUUGGGCAUUUAUCGAUGCCUAGUUAUAGAUACGAGCUU ..((((.(((((.((((.((((((((...(((........)))...))).))))).))))(((((((((....)))))))))....)).)))..)))) ( -27.40) >DroYak_CAF1 9347 98 - 1 UCAAGUAGUACUUGUACCUCUCGAAUAAAUACGGUGAGAAGUAGUAAUUACGAUAUGCACUUGGGCAUUUAUCGAUGCCUAGAUAUAGAUACGAGCCU ....((((((.((((((.(((((...........))))).))).))).))).........(((((((((....))))))))).......)))...... ( -18.80) >consensus UCAAGUAGUACUUGUACCUCUCGAAUAAAUACGGCGAGAAGUAGUAAUUACGCUAUGCACUUGGGCAUUUAUCGAUGCCCAGAUAUAGAUACGAGCUU ....((((((.((((((.(((((...........))))).))).))).))).........(((((((((....))))))))).......)))...... (-19.14 = -19.10 + -0.04)


| Location | 4,175,778 – 4,175,885 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 94.77 |
| Mean single sequence MFE | -28.02 |
| Consensus MFE | -23.92 |
| Energy contribution | -24.40 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969065 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
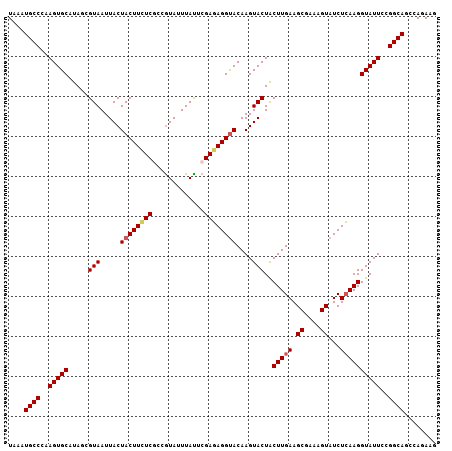
>X_DroMel_CAF1 4175778 107 + 22224390 UAAAUGCCCAAGUGCAUAGAGUAAUUACUACUUUUCGCCAUAUUUAUUCGAGAGGUACAAGUACUACUUGAAGCGAAAGUAUCUCAAGGUAUUCCGGCAGCCAGAAG ....((((..(((((...(((....((((....(((((........((((((.((((....)))).))))))))))))))).)))...)))))..))))........ ( -27.50) >DroSec_CAF1 8491 107 + 1 UAAAUGCCCAAGUGCAUAGCGUAAUUACUACUUCUCGCCGUAUUUAUACGAGAGGUACAAGUACUACUUGAAGCGAAAGUAUCUCAAGGUAUUCCGGCAGCCAGAAG ....((((..(((((.....(((.....(((((((((...........)))))))))....)))..(((((.((....))...))))))))))..))))........ ( -29.30) >DroSim_CAF1 8891 107 + 1 UAAAUGCCCAAGUGCAUAGCGUAAUUACUACUUCUCGCCGUAUUUAUACGAGAGGUACAAGUACUACUUGAAGCGAAAGUAUCUCAAGGUAUUCCGGCAGCCAGAAG ....((((..(((((.....(((.....(((((((((...........)))))))))....)))..(((((.((....))...))))))))))..))))........ ( -29.30) >DroEre_CAF1 8478 107 + 1 UAAAUGCCCAAGUGCAUAUCGUAAUUACUACUUCUCACCGUAUUUACUCGAUAGGCACAAGUACUGCUUGAAGCGAAAGUAUCUGAAGGUAUUCUGGCAGCCAGAAG ....((((...((((.((((((((.(((...........))).))))..)))).)))).((((((....((.((....)).))....))))))..))))........ ( -26.70) >DroYak_CAF1 9374 107 + 1 UAAAUGCCCAAGUGCAUAUCGUAAUUACUACUUCUCACCGUAUUUAUUCGAGAGGUACAAGUACUACUUGAAGCGAAAGUAUCUCAAGGUAUUCUGGCAGCCAGAAG ....((((..(((((.....(((.....((((((((.............))))))))....)))..(((((.((....))...))))))))))..))))........ ( -27.32) >consensus UAAAUGCCCAAGUGCAUAGCGUAAUUACUACUUCUCGCCGUAUUUAUUCGAGAGGUACAAGUACUACUUGAAGCGAAAGUAUCUCAAGGUAUUCCGGCAGCCAGAAG ....((((..(((((.....(((.....((((((((.............))))))))....)))..(((((.((....))...))))))))))..))))........ (-23.92 = -24.40 + 0.48)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:08:08 2006