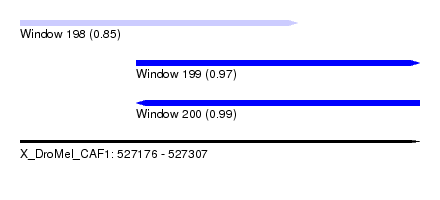
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 527,176 – 527,307 |
| Length | 131 |
| Max. P | 0.990698 |

| Location | 527,176 – 527,267 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 85.28 |
| Mean single sequence MFE | -28.28 |
| Consensus MFE | -20.14 |
| Energy contribution | -21.33 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.850997 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 527176 91 + 22224390 UAUAUCGCCUUUCAGUAUGGACAG--GAACAGCGGCUAUAUCCUU-------------CGGUAUCCCCCUCGCCACAUUCGCCAUAUGCGAGUGUGGCGUACAUAA .......(((.((......)).))--)...((.((..(((((...-------------.)))))..))))((((((((((((.....))))))))))))....... ( -28.80) >DroSec_CAF1 13758 91 + 1 UAUAUCGCCUUUCAGUAUGGACAG--GAACAGCGGCUAUAUCCUU-------------CGGUAUCCCCCUCGCCACAUUCGCCAUAUGCGAGUGUGGCGUACAUAA .......(((.((......)).))--)...((.((..(((((...-------------.)))))..))))((((((((((((.....))))))))))))....... ( -28.80) >DroSim_CAF1 14071 91 + 1 UAUAUCGCCUUUCAGUAUGGACAG--GAACAGCGGCUAUAUCCUU-------------CGGUAUCCCCCUCGCCACAUUCGCCAUAUGCGAGUGUGGCGUACAUAA .......(((.((......)).))--)...((.((..(((((...-------------.)))))..))))((((((((((((.....))))))))))))....... ( -28.80) >DroEre_CAF1 18652 100 + 1 UAUAUCGCCUUUCAGUAUGGACAGCGGAACGGCGGCCAUAUCCUACUACCACCACGUA-GAUAUACCCCUCG-----GUCGCCAUAUUUGAGUGUGGCGUACAUAA .....((((.((((((((((.(.((.((..((.((..(((((.(((.........)))-))))).)))))).-----)).)))))).)))))...))))....... ( -26.70) >consensus UAUAUCGCCUUUCAGUAUGGACAG__GAACAGCGGCUAUAUCCUU_____________CGGUAUCCCCCUCGCCACAUUCGCCAUAUGCGAGUGUGGCGUACAUAA ...............((((.((........((.((..(((((.................)))))..)))).(((((((((((.....))))))))))))).)))). (-20.14 = -21.33 + 1.19)

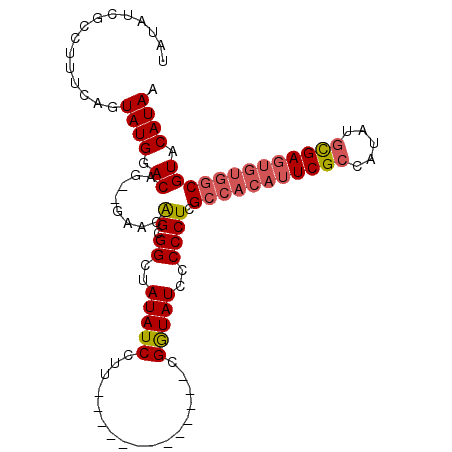
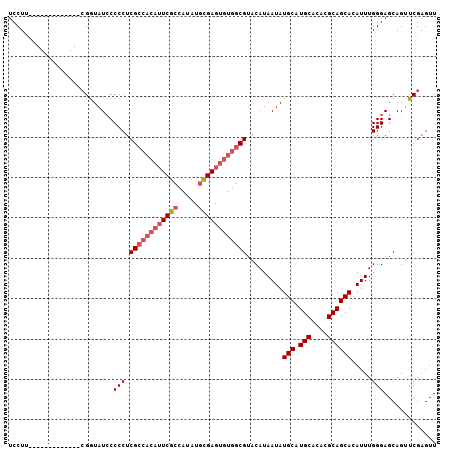
| Location | 527,214 – 527,307 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 87.44 |
| Mean single sequence MFE | -33.48 |
| Consensus MFE | -25.06 |
| Energy contribution | -26.62 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.97 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.971014 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 527214 93 + 22224390 UCCUU-------------CGGUAUCCCCCUCGCCACAUUCGCCAUAUGCGAGUGUGGCGUACAUAAUAUGCAUGCACACGCAGCACAUUUGGGAGCAGUUCGAGUU ...((-------------(((..((((...((((((((((((.....)))))))))))).........(((.(((....)))))).....)))).....))))).. ( -36.10) >DroSec_CAF1 13796 93 + 1 UCCUU-------------CGGUAUCCCCCUCGCCACAUUCGCCAUAUGCGAGUGUGGCGUACAUAAUAUGCAUGCACACGCAGCACAUUUGGGAGCAGUUCGAGUU ...((-------------(((..((((...((((((((((((.....)))))))))))).........(((.(((....)))))).....)))).....))))).. ( -36.10) >DroSim_CAF1 14109 93 + 1 UCCUU-------------CGGUAUCCCCCUCGCCACAUUCGCCAUAUGCGAGUGUGGCGUACAUAAUAUGCAUGCACACGCAGCACAUUUGGGAGCAGUUCGAGUU ...((-------------(((..((((...((((((((((((.....)))))))))))).........(((.(((....)))))).....)))).....))))).. ( -36.10) >DroEre_CAF1 18692 100 + 1 UCCUACUACCACCACGUA-GAUAUACCCCUCG-----GUCGCCAUAUUUGAGUGUGGCGUACAUAAUAUGCAUGCACACGCAGCACAUUUGGGAGCAGUUCGAGUU .....((((......)))-)........((((-----..((((((((....)))))))((.(..(((.(((.(((....)))))).)))...).)).)..)))).. ( -25.60) >consensus UCCUU_____________CGGUAUCCCCCUCGCCACAUUCGCCAUAUGCGAGUGUGGCGUACAUAAUAUGCAUGCACACGCAGCACAUUUGGGAGCAGUUCGAGUU ..........................(((.((((((((((((.....)))))))))))).........(((.(((....)))))).....)))............. (-25.06 = -26.62 + 1.56)

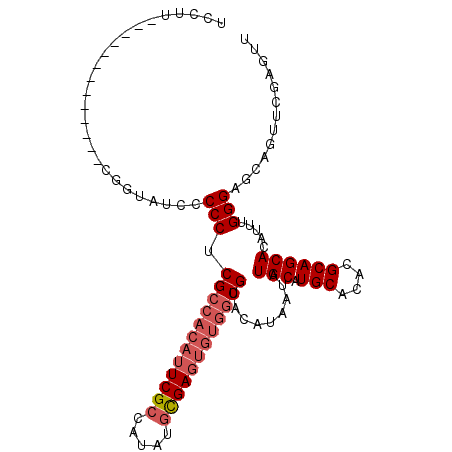
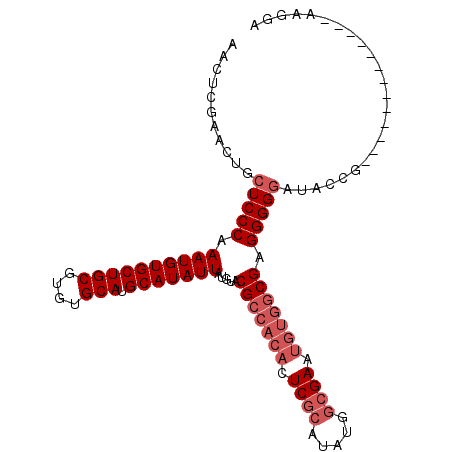
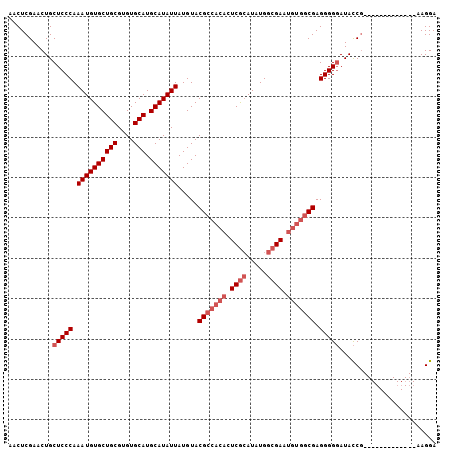
| Location | 527,214 – 527,307 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 87.44 |
| Mean single sequence MFE | -37.67 |
| Consensus MFE | -29.85 |
| Energy contribution | -31.85 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.40 |
| Structure conservation index | 0.79 |
| SVM decision value | 2.23 |
| SVM RNA-class probability | 0.990698 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 527214 93 - 22224390 AACUCGAACUGCUCCCAAAUGUGCUGCGUGUGCAUGCAUAUUAUGUACGCCACACUCGCAUAUGGCGAAUGUGGCGAGGGGGAUACCG-------------AAGGA ...(((.....(((((.((((((((((....))).))))))).....(((((((.((((.....)))).))))))).)))))....))-------------).... ( -40.00) >DroSec_CAF1 13796 93 - 1 AACUCGAACUGCUCCCAAAUGUGCUGCGUGUGCAUGCAUAUUAUGUACGCCACACUCGCAUAUGGCGAAUGUGGCGAGGGGGAUACCG-------------AAGGA ...(((.....(((((.((((((((((....))).))))))).....(((((((.((((.....)))).))))))).)))))....))-------------).... ( -40.00) >DroSim_CAF1 14109 93 - 1 AACUCGAACUGCUCCCAAAUGUGCUGCGUGUGCAUGCAUAUUAUGUACGCCACACUCGCAUAUGGCGAAUGUGGCGAGGGGGAUACCG-------------AAGGA ...(((.....(((((.((((((((((....))).))))))).....(((((((.((((.....)))).))))))).)))))....))-------------).... ( -40.00) >DroEre_CAF1 18692 100 - 1 AACUCGAACUGCUCCCAAAUGUGCUGCGUGUGCAUGCAUAUUAUGUACGCCACACUCAAAUAUGGCGAC-----CGAGGGGUAUAUC-UACGUGGUGGUAGUAGGA .......(((((..((.((((((((((....))).)))))))((((((((((..........)))))((-----(....))).....-)))))))..))))).... ( -30.70) >consensus AACUCGAACUGCUCCCAAAUGUGCUGCGUGUGCAUGCAUAUUAUGUACGCCACACUCGCAUAUGGCGAAUGUGGCGAGGGGGAUACCG_____________AAGGA ...........(((((.((((((((((....))).))))))).....(((((((.((((.....)))).))))))).)))))........................ (-29.85 = -31.85 + 2.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:36:01 2006