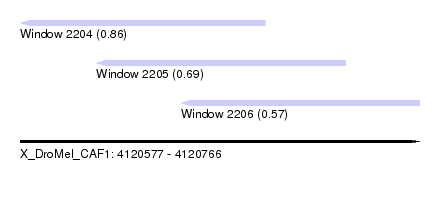
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,120,577 – 4,120,766 |
| Length | 189 |
| Max. P | 0.855598 |

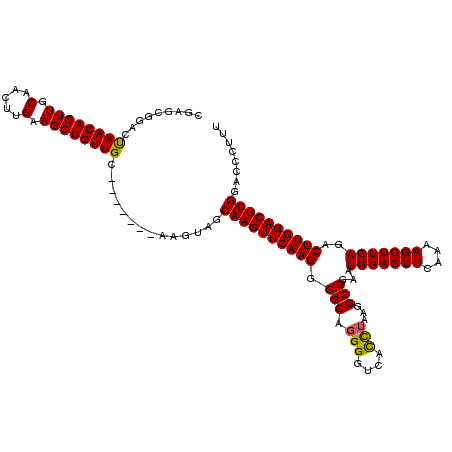
| Location | 4,120,577 – 4,120,693 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.82 |
| Mean single sequence MFE | -29.97 |
| Consensus MFE | -28.27 |
| Energy contribution | -27.83 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.855598 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4120577 116 - 22224390 AAUGGAGUUCAAAAGCUUCAGACUUGGACUUGGACCCUUUAACCUGGGACCCACAAGCCAGGAAAAGGAGCCAACACCCGACACUUGUAAAUUUCUUAUGAGCACCAAUGCAAAAA---- ..(((.(((((.(((......((((((..((((..(((((..(((((..........))))).)))))..))))...)))).....))......))).))))).))).........---- ( -29.10) >DroSim_CAF1 7490 120 - 1 AAUGGAGUUCAAAAGCUUCAGACUUGGACUUGGGCCCUUUAACCUGGGACCCACAAGCCAGGAAAAGGAGCCAACACCCGACACUUGUAAAUUUCUUAUGAGCACCAAUGCGAAAAACAA ..(((.(((((...(((((...(((((.(((((((((........))).))))..)))))))....))))).........((....))..........))))).)))............. ( -32.10) >DroYak_CAF1 8692 115 - 1 AAUGGAGUUCAAGAGCUUCAGACUUGGACUUGGAACCUUUAACCUGGGACUCACAAGCCAGGAAAAGGAGCCAACACCUGACACUUGUAAAUUUCUUAUGAGCACCAAUGCAAAA----- ..(((((((....)))))))(..((((..((((..(((((..(((((..........))))).)))))..)))).........((..(((.....)))..))..))))..)....----- ( -28.70) >consensus AAUGGAGUUCAAAAGCUUCAGACUUGGACUUGGACCCUUUAACCUGGGACCCACAAGCCAGGAAAAGGAGCCAACACCCGACACUUGUAAAUUUCUUAUGAGCACCAAUGCAAAAA____ ..(((.(((((.(((......((((((..((((..(((((..(((((..........))))).)))))..))))...)))).....))......))).))))).)))............. (-28.27 = -27.83 + -0.44)

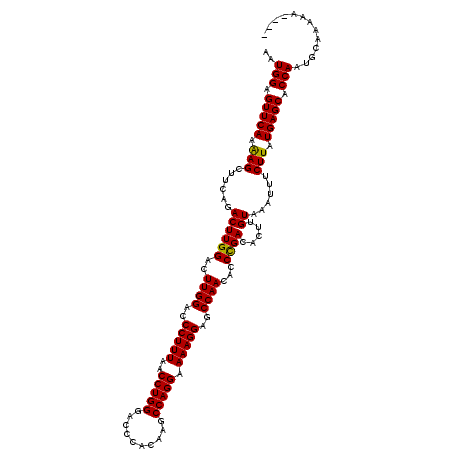
| Location | 4,120,613 – 4,120,731 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.18 |
| Mean single sequence MFE | -39.67 |
| Consensus MFE | -34.81 |
| Energy contribution | -34.70 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.690609 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4120613 118 - 22224390 --AAGUAGCAAGUUCAAGGGGCAGGGGUCACCUAAGGCUGAAUGGAGUUCAAAAGCUUCAGACUUGGACUUGGACCCUUUAACCUGGGACCCACAAGCCAGGAAAAGGAGCCAACACCCG --.......(((((((((.(((.(((....)))...)))...((((((......))))))..)))))))))((..(((((..(((((..........))))).)))))..))........ ( -37.00) >DroSim_CAF1 7530 118 - 1 --AAGUAGCAAGUUCAAGGGGCAGGGGUUACCUAAGGCUGAAUGGAGUUCAAAAGCUUCAGACUUGGACUUGGGCCCUUUAACCUGGGACCCACAAGCCAGGAAAAGGAGCCAACACCCG --......((((((((((.(((.(((....)))...)))...((((((......))))))..))))))))))((((((((..(((((..........))))).))))).)))........ ( -42.80) >DroYak_CAF1 8727 120 - 1 GCAAGUAGCAAGUUCAAGGGGCAGGGGCCAUUGCUGGCUGAAUGGAGUUCAAGAGCUUCAGACUUGGACUUGGAACCUUUAACCUGGGACUCACAAGCCAGGAAAAGGAGCCAACACCUG ((......((((((((((.......(((((....)))))...(((((((....)))))))..))))))))))...(((((..(((((..........))))).))))).))......... ( -39.20) >consensus __AAGUAGCAAGUUCAAGGGGCAGGGGUCACCUAAGGCUGAAUGGAGUUCAAAAGCUUCAGACUUGGACUUGGACCCUUUAACCUGGGACCCACAAGCCAGGAAAAGGAGCCAACACCCG .........(((((((((.(((.(((....)))...)))...(((((((....)))))))..)))))))))((..(((((..(((((..........))))).)))))..))........ (-34.81 = -34.70 + -0.11)


| Location | 4,120,653 – 4,120,766 |
|---|---|
| Length | 113 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.97 |
| Mean single sequence MFE | -38.30 |
| Consensus MFE | -30.83 |
| Energy contribution | -30.50 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.571302 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4120653 113 - 22224390 CAAGCGGACUAACAGUUGCAACUUGAAGCUGUUGC-------AAGUAGCAAGUUCAAGGGGCAGGGGUCACCUAAGGCUGAAUGGAGUUCAAAAGCUUCAGACUUGGACUUGGACCCUUU .(((.((......(.(((((((........)))))-------)).)..((((((((((.(((.(((....)))...)))...((((((......))))))..))))))))))..))))). ( -36.80) >DroSim_CAF1 7570 111 - 1 CGAGCGGACCAACAGUUGCA--CUGAAGCUGUUGC-------AAGUAGCAAGUUCAAGGGGCAGGGGUUACCUAAGGCUGAAUGGAGUUCAAAAGCUUCAGACUUGGACUUGGGCCCUUU .(((.((.(((((((((...--....)))))))..-------......((((((((((.(((.(((....)))...)))...((((((......))))))..))))))))))))))))). ( -37.70) >DroYak_CAF1 8767 120 - 1 CGAGCUGACUAACAGUUGCAACUUGAAGCUGUUGCAAGUUGCAAGUAGCAAGUUCAAGGGGCAGGGGCCAUUGCUGGCUGAAUGGAGUUCAAGAGCUUCAGACUUGGACUUGGAACCUUU .(((((..(((....((((((((((.........)))))))))).)))..)))))((((..(...(((((....))))).....(((((((((.........))))))))).)..)))). ( -40.40) >consensus CGAGCGGACUAACAGUUGCAACUUGAAGCUGUUGC_______AAGUAGCAAGUUCAAGGGGCAGGGGUCACCUAAGGCUGAAUGGAGUUCAAAAGCUUCAGACUUGGACUUGGACCCUUU .........((((((((.(.....).))))))))..............((((((((((.(((.(((....)))...)))...(((((((....)))))))..))))))))))........ (-30.83 = -30.50 + -0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:07:58 2006