
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,118,619 – 4,118,844 |
| Length | 225 |
| Max. P | 0.857849 |

| Location | 4,118,619 – 4,118,725 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.99 |
| Mean single sequence MFE | -36.27 |
| Consensus MFE | -28.92 |
| Energy contribution | -29.43 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.857849 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
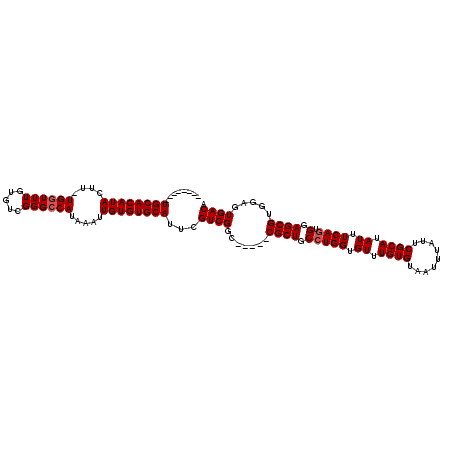
>X_DroMel_CAF1 4118619 106 + 22224390 --------UGCACAUACUU-UGGUUUGUGUCGGGCCAUAAAUUGUGUGCAUUCGUCGGC-----CGCUGCCUGGUGUUUGUGUAAUUUAUUCGCAUAUUUCAGUGGAGCGUGGAGUGACA --------((((((((...-(((((((...))))))).....))))))))...((((.(-----(((.(((((.((..((((..(.....)..))))...)).))).))))))..)))). ( -34.30) >DroSec_CAF1 9710 111 + 1 --------UGCACAUACUU-UGGUUUGUGUCGGGACAUAAAUUGUGUGCAUUCGUCGGCUGGUCCGCUGCCUGGUGUUUGUGUAAUUUAUUCGCAUAUUUCAGUGGAGCGUGGAGUGACA --------((((((((...-..((((((((....)))))))))))))))).(((((.((.(.(((((((....((((..(((.....)))..))))....))))))).))).)).))).. ( -35.90) >DroSim_CAF1 5480 120 + 1 CGGUUAAUUGCACAUACUUNUGGUUUGUGUCGGGCCAUAAAUUGUGUGCAUUCGUCGGCUGGUCCGCUGCCUGGUGUUUGUGUAAUUUAUUCGCAUAUUUCAGUGGAGCGUGGAGUGACA ..((((..((((((((....(((((((...))))))).....))))))))....((.((.(.(((((((....((((..(((.....)))..))))....))))))).))).)).)))). ( -37.60) >DroYak_CAF1 6253 106 + 1 --------UGCACAUACUU-UGGUUUGUGUUGGGCCAUAAAUUGUGUGCAUUCGUCGGU-----CGCUGCCUGGUGUUUGUGUAAUUUAUUCGCAUAUUUCACUGGAGCGCGGAGUGACA --------((((((((...-((((((.....)))))).....))))))))...((((.(-----(((.(((..(((..((((..(.....)..))))...)))..).))))))..)))). ( -37.30) >consensus ________UGCACAUACUU_UGGUUUGUGUCGGGCCAUAAAUUGUGUGCAUUCGUCGGC_____CGCUGCCUGGUGUUUGUGUAAUUUAUUCGCAUAUUUCAGUGGAGCGUGGAGUGACA ........((((((((....((((((.....)))))).....))))))))...((((.......((((.(((((.((.((((.........)))).)).)))).).)))).....)))). (-28.92 = -29.43 + 0.50)

| Location | 4,118,725 – 4,118,844 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.44 |
| Mean single sequence MFE | -35.58 |
| Consensus MFE | -30.61 |
| Energy contribution | -31.99 |
| Covariance contribution | 1.38 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.655133 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4118725 119 - 22224390 UGCCCCUGCUGGAUGUCUGCCCUCGUUUCUGUCUUGACUUG-GGCCACAUCUAUUACGCCCGCUCCCCAAAAACUUAACGGGGGUAGGCAUUCCUGGCAUUUUGUAGCAUAAUUUCAGGC ....((((..(((((((((((((((((...((.(((...((-(((............))))).....)))..))..)))))))))))))))))...((........)).......)))). ( -38.60) >DroSec_CAF1 9821 120 - 1 UGCCCCUGCUGGAUGUCUGCCCUCGUUUCUGUCUUGACUUGGGGCCACAUCUAUUACGCCCGCUCCUCAAAAACUUAACAGGGGUAGGCAUUCCUGGCAUUUUGUAGCAUAAUUUCAGGC ....((((..((((((((((((..(((...((.((((...(((((............)))).)...))))..))..)))..))))))))))))...((........)).......)))). ( -36.20) >DroSim_CAF1 5600 120 - 1 UGCCCCUGCUGGAUGUCUGCCCUCGUUUCUGUCUUGACUUGGGGCCACAUCUAUUACGCCCGCUCCCCAAAAACUUAACGGGGGUAGGCAUUCCUGGCAUUUUGUAGCAUAAUUUCAGGC ....((((..(((((((((((((((((...((.(((....(((((............)))).)....)))..))..)))))))))))))))))...((........)).......)))). ( -39.10) >DroYak_CAF1 6359 112 - 1 UGCCCCUUUUGGAUGUCUGCCCUUGUUUCUGUCUUGACUUG--------UCUAUUACGCCCGCUCCCCAAAAACUUAACGGGGGUAGGCAUUCCUGGCAUUUUGUAGCAUAAUUUCAGGC .(((......(((((((((((((((((...((.(((....(--------(...........))....)))..))..)))))))))))))))))...((........)).........))) ( -28.40) >consensus UGCCCCUGCUGGAUGUCUGCCCUCGUUUCUGUCUUGACUUG_GGCCACAUCUAUUACGCCCGCUCCCCAAAAACUUAACGGGGGUAGGCAUUCCUGGCAUUUUGUAGCAUAAUUUCAGGC .(((......(((((((((((((((((...((.((.....(((((................)))))...)).))..)))))))))))))))))...((........)).........))) (-30.61 = -31.99 + 1.38)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:07:53 2006