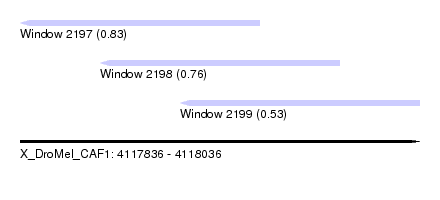
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,117,836 – 4,118,036 |
| Length | 200 |
| Max. P | 0.826940 |

| Location | 4,117,836 – 4,117,956 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.69 |
| Mean single sequence MFE | -25.38 |
| Consensus MFE | -21.35 |
| Energy contribution | -20.42 |
| Covariance contribution | -0.94 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.826940 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4117836 120 - 22224390 CUUCAUGCAAUGUCAUGACAAAUGACACUCAGCUCAUCCUAGGAGCAGCAAUGAAACUUGCUGAAUUGAAGCUCUUCUCCUACACAGUGAACAACAAGCAGUUCAUUAACAUACUAUAGG .(((((((..((((((.....))))))....((((.......)))).)).)))))....(((.......))).............(((((((........)))))))............. ( -25.40) >DroSec_CAF1 8955 120 - 1 CUUCAUGCAAUGUCAUGACAAAUGACACUCAGCUCAUCCUAGGAGUAGCAAUGAAACUUGCUGAAUUGAAGCUCUUCCCUUACACAGUGAACAACAGGCAGUUCAUUAACAUACUAUAAG (((((.((((((((((.....))))))....(((..((....))..)))........)))).....)))))..............(((((((........)))))))............. ( -23.50) >DroSim_CAF1 4359 120 - 1 CUUCAUGCAAUGUCAUGACAAAUGACACUCAGCUCAUCCUAGGAGCAGCAAUGAAACUUGCUGAAUUGAAGCUCUUCCCUUACACAGUGAACAACAAGCAGUUCAUUAACAUACUAUAAG .(((((((..((((((.....))))))....((((.......)))).)).)))))....(((.......))).............(((((((........)))))))............. ( -25.40) >DroYak_CAF1 4641 113 - 1 AUUCAUGCAAUGUCAUGACAAAUGACACUCAGCUCAUCCUGGGAGCCGC-AUGAAACUUGGUGAAUUGAAGCUCUUCUCUUACACAGCGAAAA--AAUCAGUUAAUU----UAUUAUAAA .(((((((..((((((.....))))))(((((......)))))....))-)))))...(((((((((((.(((............)))((...--..))..))))))----))))).... ( -27.20) >consensus CUUCAUGCAAUGUCAUGACAAAUGACACUCAGCUCAUCCUAGGAGCAGCAAUGAAACUUGCUGAAUUGAAGCUCUUCCCUUACACAGUGAACAACAAGCAGUUCAUUAACAUACUAUAAG .(((((((..((((((.....))))))....((((.......)))).)).)))))..(((((........(((............)))........)))))................... (-21.35 = -20.42 + -0.94)



| Location | 4,117,876 – 4,117,996 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.69 |
| Mean single sequence MFE | -31.95 |
| Consensus MFE | -27.89 |
| Energy contribution | -28.45 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.762231 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4117876 120 - 22224390 UCCGUUCGCACACAUGCCUCGCCGCUUGGUUCACGCUUCUCUUCAUGCAAUGUCAUGACAAAUGACACUCAGCUCAUCCUAGGAGCAGCAAUGAAACUUGCUGAAUUGAAGCUCUUCUCC ..((...(((....)))..))..(((..(((((.((.....(((((((..((((((.....))))))....((((.......)))).)).)))))....)))))))..).))........ ( -30.00) >DroSec_CAF1 8995 120 - 1 UCCGUUCGCACACAUGCCUCGGCGCUUGGUUCACGCUUCUCUUCAUGCAAUGUCAUGACAAAUGACACUCAGCUCAUCCUAGGAGUAGCAAUGAAACUUGCUGAAUUGAAGCUCUUCCCU ...((..((.((..(((....)))..))))..))(((((..((((.((((((((((.....))))))....(((..((....))..)))........))))))))..)))))........ ( -31.40) >DroSim_CAF1 4399 120 - 1 UCCGUUCGCACACAUGCCUCGGCGCUUGGUUCACGCUUCUCUUCAUGCAAUGUCAUGACAAAUGACACUCAGCUCAUCCUAGGAGCAGCAAUGAAACUUGCUGAAUUGAAGCUCUUCCCU .(((...(((....)))..))).(((..(((((.((.....(((((((..((((((.....))))))....((((.......)))).)).)))))....)))))))..).))........ ( -33.10) >DroYak_CAF1 4675 119 - 1 UCCGUUCGCACACAUGCCUCUGCGCUUGGUUCACGCUUCUAUUCAUGCAAUGUCAUGACAAAUGACACUCAGCUCAUCCUGGGAGCCGC-AUGAAACUUGGUGAAUUGAAGCUCUUCUCU ...((..(((....)))....))(((..((((((.(.....(((((((..((((((.....))))))(((((......)))))....))-)))))....)))))))..).))........ ( -33.30) >consensus UCCGUUCGCACACAUGCCUCGGCGCUUGGUUCACGCUUCUCUUCAUGCAAUGUCAUGACAAAUGACACUCAGCUCAUCCUAGGAGCAGCAAUGAAACUUGCUGAAUUGAAGCUCUUCCCU .(((...(((....)))..))).(((..(((((.((.....(((((((..((((((.....))))))....((((.......)))).)).)))))....)))))))..).))........ (-27.89 = -28.45 + 0.56)

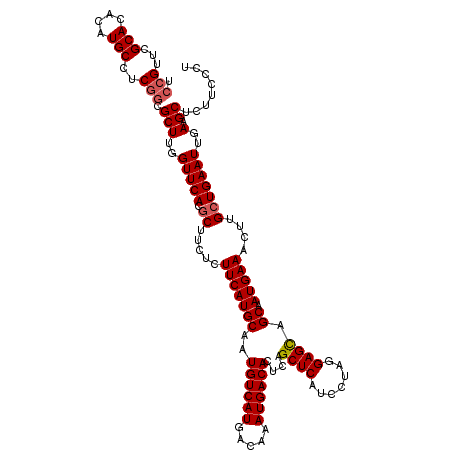

| Location | 4,117,916 – 4,118,036 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.36 |
| Mean single sequence MFE | -26.38 |
| Consensus MFE | -23.35 |
| Energy contribution | -24.10 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.89 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.528872 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4117916 120 - 22224390 GAUCAAGCUGGCUGUGAACUAUUUAACAUUUAAACUCAACUCCGUUCGCACACAUGCCUCGCCGCUUGGUUCACGCUUCUCUUCAUGCAAUGUCAUGACAAAUGACACUCAGCUCAUCCU (((..(((((((.((((((((...........(((........))).(((....))).........))))))))))..............((((((.....))))))..))))).))).. ( -24.80) >DroSec_CAF1 9035 120 - 1 GAUCAAGCUGGCUGUGAACUAUUUAACAUUUAAACCCAACUCCGUUCGCACACAUGCCUCGGCGCUUGGUUCACGCUUCUCUUCAUGCAAUGUCAUGACAAAUGACACUCAGCUCAUCCU (((..(((((((.((((((((((((.....)))).....(.(((...(((....)))..))).)..))))))))))..............((((((.....))))))..))))).))).. ( -28.00) >DroSim_CAF1 4439 120 - 1 GAUCAAGCUGGCUGUGAACUAUUUAACAUUUAAACCCAACUCCGUUCGCACACAUGCCUCGGCGCUUGGUUCACGCUUCUCUUCAUGCAAUGUCAUGACAAAUGACACUCAGCUCAUCCU (((..(((((((.((((((((((((.....)))).....(.(((...(((....)))..))).)..))))))))))..............((((((.....))))))..))))).))).. ( -28.00) >DroYak_CAF1 4714 120 - 1 GAUCAAGAUGGCAGUGAACUAUUUAACAUUUAAACUCAACUCCGUUCGCACACAUGCCUCUGCGCUUGGUUCACGCUUCUAUUCAUGCAAUGUCAUGACAAAUGACACUCAGCUCAUCCU ......((((((.((((((((...........(((........)))((((..........))))..))))))))((..........))..((((((.....))))))....)).)))).. ( -24.70) >consensus GAUCAAGCUGGCUGUGAACUAUUUAACAUUUAAACCCAACUCCGUUCGCACACAUGCCUCGGCGCUUGGUUCACGCUUCUCUUCAUGCAAUGUCAUGACAAAUGACACUCAGCUCAUCCU (((..(((((((.((((((((((((.....)))).....(.(((...(((....)))..))).)..))))))))))..............((((((.....))))))..))))).))).. (-23.35 = -24.10 + 0.75)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:07:51 2006