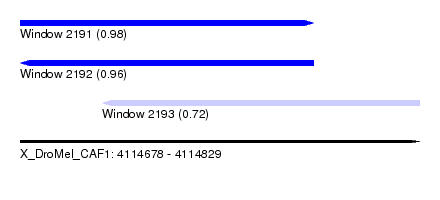
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,114,678 – 4,114,829 |
| Length | 151 |
| Max. P | 0.982750 |

| Location | 4,114,678 – 4,114,789 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 92.19 |
| Mean single sequence MFE | -45.07 |
| Consensus MFE | -41.29 |
| Energy contribution | -41.40 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.19 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.92 |
| SVM RNA-class probability | 0.982750 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4114678 111 + 22224390 AGCACCAACACGUGCAGCGAUCAGGGGCAAGGACAUGGAUCUGGGAUUGGACUUGGUGACCCAAAGCGGGACCAAAGCCAAGUCCACUACGCCCAUGACCCUGCCCAGGAU .((((......))))....(((..(((((.((.(((((...(((...((((((((((..(((.....)))......)))))))))))))...))))).)).)))))..))) ( -47.50) >DroSec_CAF1 6017 111 + 1 AGCACCAACACGUGCAGCGAUCAGGGGCAAGGACAUGGAUCUGGGAUUGGACUUGGUGACCCAAAGCGGGACCAAAGCCAAGUCCACUACGCCCAUGACCCUGCCCAGGAU .((((......))))....(((..(((((.((.(((((...(((...((((((((((..(((.....)))......)))))))))))))...))))).)).)))))..))) ( -47.50) >DroYak_CAF1 1357 102 + 1 ---------ACGUGCAGCGAUCAGGGGCAAGGACAUGGAUCUGGCAUUGGGCUUGGUGACCCAAAGCGGACCCAAAGCCAAGUCCACUACGCCCAUGACCCUGCCCAGGAU ---------..(.((((.(.(((.((((..((...((((..((((.(((((.((.((........)).))))))).))))..))))))..)))).))).))))).)..... ( -40.20) >consensus AGCACCAACACGUGCAGCGAUCAGGGGCAAGGACAUGGAUCUGGGAUUGGACUUGGUGACCCAAAGCGGGACCAAAGCCAAGUCCACUACGCCCAUGACCCUGCCCAGGAU ..........((.....))(((..(((((.((.(((((...(((...((((((((((..(((.....)))......)))))))))))))...))))).)).)))))..))) (-41.29 = -41.40 + 0.11)

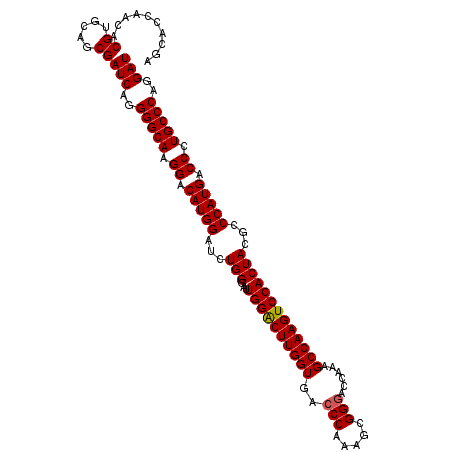
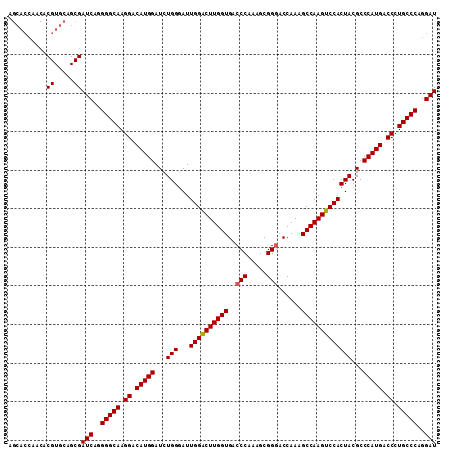
| Location | 4,114,678 – 4,114,789 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 92.19 |
| Mean single sequence MFE | -51.07 |
| Consensus MFE | -43.49 |
| Energy contribution | -44.93 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.84 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.955381 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4114678 111 - 22224390 AUCCUGGGCAGGGUCAUGGGCGUAGUGGACUUGGCUUUGGUCCCGCUUUGGGUCACCAAGUCCAAUCCCAGAUCCAUGUCCUUGCCCCUGAUCGCUGCACGUGUUGGUGCU (((..((((((((.(((((((....(((((((((...((..((......))..)))))))))))......).)))))).))))))))..)))....((((......)))). ( -52.00) >DroSec_CAF1 6017 111 - 1 AUCCUGGGCAGGGUCAUGGGCGUAGUGGACUUGGCUUUGGUCCCGCUUUGGGUCACCAAGUCCAAUCCCAGAUCCAUGUCCUUGCCCCUGAUCGCUGCACGUGUUGGUGCU (((..((((((((.(((((((....(((((((((...((..((......))..)))))))))))......).)))))).))))))))..)))....((((......)))). ( -52.00) >DroYak_CAF1 1357 102 - 1 AUCCUGGGCAGGGUCAUGGGCGUAGUGGACUUGGCUUUGGGUCCGCUUUGGGUCACCAAGCCCAAUGCCAGAUCCAUGUCCUUGCCCCUGAUCGCUGCACGU--------- .......(((((((((.(((((..(((((.(((((.((((((.....((((....)))))))))).))))).))))).....))))).))))).))))....--------- ( -49.20) >consensus AUCCUGGGCAGGGUCAUGGGCGUAGUGGACUUGGCUUUGGUCCCGCUUUGGGUCACCAAGUCCAAUCCCAGAUCCAUGUCCUUGCCCCUGAUCGCUGCACGUGUUGGUGCU (((..((((((((.(((((((....(((((((((...((..((......))..)))))))))))......).)))))).))))))))..)))....((((......)))). (-43.49 = -44.93 + 1.45)

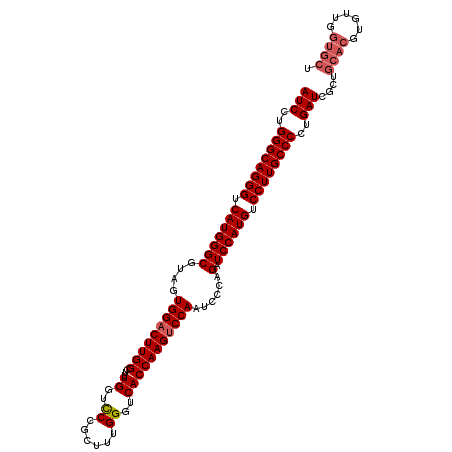
| Location | 4,114,709 – 4,114,829 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -41.62 |
| Consensus MFE | -36.69 |
| Energy contribution | -37.00 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.724188 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4114709 120 - 22224390 ACAUUCCAGUACAACUCUUGACAUUCGGUGAAUCGCCAUUAUCCUGGGCAGGGUCAUGGGCGUAGUGGACUUGGCUUUGGUCCCGCUUUGGGUCACCAAGUCCAAUCCCAGAUCCAUGUC .....((((..((.....))......((((...))))......))))((((((((.((((.....(((((((((...((..((......))..)))))))))))..))))))))).))). ( -41.90) >DroSec_CAF1 6048 120 - 1 ACAUUCCAGUACAACUCCUGACAUUCGGUGAAUCGCCAUUAUCCUGGGCAGGGUCAUGGGCGUAGUGGACUUGGCUUUGGUCCCGCUUUGGGUCACCAAGUCCAAUCCCAGAUCCAUGUC .....((((..((.....))......((((...))))......))))((((((((.((((.....(((((((((...((..((......))..)))))))))))..))))))))).))). ( -42.10) >DroSim_CAF1 1496 120 - 1 ACAUUCCAGUACAACUCCUGACAUUCGGUGAAUCGCCAUUAUCCUGGGCAGGGUCAUGGGCGUAGUGGACUUGGCUUUGGUCCCGCUUUGGGUCACCAAGUCCAAUCCCAGAUCCAUGUC .....((((..((.....))......((((...))))......))))((((((((.((((.....(((((((((...((..((......))..)))))))))))..))))))))).))). ( -42.10) >DroYak_CAF1 1379 120 - 1 UCAAUUCAUUACAACUCUUGACAUUCGGUGAAUCGCCAUUAUCCUGGGCAGGGUCAUGGGCGUAGUGGACUUGGCUUUGGGUCCGCUUUGGGUCACCAAGCCCAAUGCCAGAUCCAUGUC .........(((..(((.((((....((((...)))).....(((....))))))).))).)))(((((.(((((.((((((.....((((....)))))))))).))))).)))))... ( -40.40) >consensus ACAUUCCAGUACAACUCCUGACAUUCGGUGAAUCGCCAUUAUCCUGGGCAGGGUCAUGGGCGUAGUGGACUUGGCUUUGGUCCCGCUUUGGGUCACCAAGUCCAAUCCCAGAUCCAUGUC ...................(((((..........(((.........))).(((((.((((.....(((((((((...((..((......))..)))))))))))..)))))))))))))) (-36.69 = -37.00 + 0.31)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:07:45 2006