
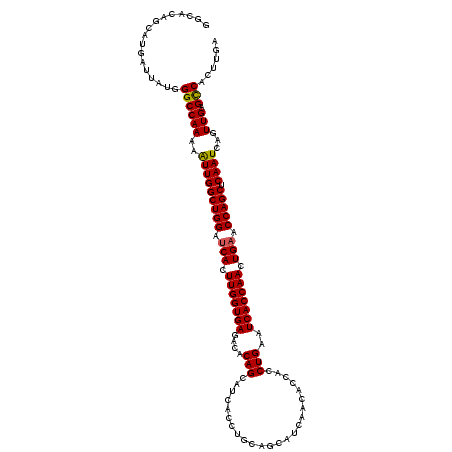
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,114,322 – 4,114,504 |
| Length | 182 |
| Max. P | 0.964056 |

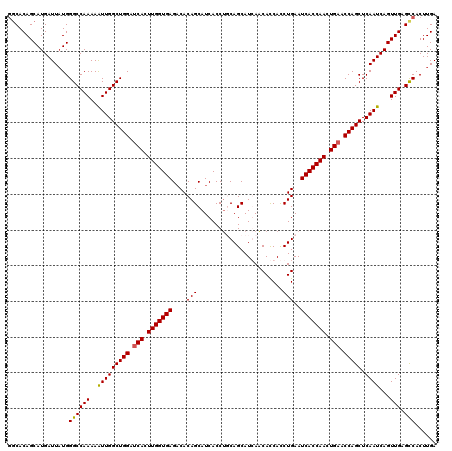
| Location | 4,114,322 – 4,114,442 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.31 |
| Mean single sequence MFE | -36.10 |
| Consensus MFE | -31.24 |
| Energy contribution | -31.11 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.964056 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4114322 120 + 22224390 GGCACAGCAUGAUUAUGGGCCAAAAGUUGGCUGGAUCACUUGGUGAGACACAGCAUCACCUUCAGCAUCAACAUCACCUGAAUCACCAACUGAACCAGCUCAAUCAGUUGAGCCACUUGA (((.((((.(((((...(((.....)))((((((.(((.(((((((......((..........)).(((........))).))))))).))).)))))).))))))))).)))...... ( -36.70) >DroSec_CAF1 5532 120 + 1 UUCACAGCAUGAUUAUGGGCCAAAAAUUGGCUGGAUCACUUGGUGAGACACAGCAUCACCUGCAGCAUGAACACCACCUGAAUCACCAACUGAACCAGCUCAAUCAGUUGAGCCACUUGA .................((((((..(((((((((.(((.(((((((..((..(((.....)))....((.....))..))..))))))).))).))))).))))...))).)))...... ( -34.60) >DroSim_CAF1 1025 120 + 1 GGCACAGCAUGAUUAUGGGCCAAAAAUUGGCUGGAUCACUUGGUGAGACACAGCAUCACCUGCAGCAUGAACACCACCUGAAUCACCAACUGAACCAGCUCAAUCAGUUGAGCCACUUGA (((.((((.(((((...(((((.....)))))((.(((.(((((((..((..(((.....)))....((.....))..))..))))))).))).)).....))))))))).)))...... ( -36.90) >DroYak_CAF1 986 120 + 1 GGCACAGCAUGAUUCUGGGCCAAAAAUUGGCUGGAGCACUUGGUGAGCCACAGCAUCACCAUCAGCAUCAUCACCAUCUGAAUCACCAACUGAACCAGCUCAAUCAGUUGAGUCACUUGA (((......(((((((((((((.....))))((..((...((((((((....)).))))))...))..))...)))...)))))).(((((((..........))))))).)))...... ( -36.20) >consensus GGCACAGCAUGAUUAUGGGCCAAAAAUUGGCUGGAUCACUUGGUGAGACACAGCAUCACCUGCAGCAUCAACACCACCUGAAUCACCAACUGAACCAGCUCAAUCAGUUGAGCCACUUGA .................((((((..(((((((((.(((.(((((((....(((........................)))..))))))).))).))))).))))...))).)))...... (-31.24 = -31.11 + -0.13)

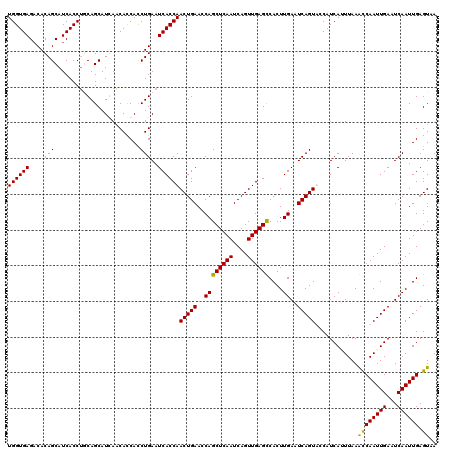
| Location | 4,114,362 – 4,114,482 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.61 |
| Mean single sequence MFE | -25.00 |
| Consensus MFE | -21.39 |
| Energy contribution | -20.76 |
| Covariance contribution | -0.63 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.876200 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4114362 120 + 22224390 UGGUGAGACACAGCAUCACCUUCAGCAUCAACAUCACCUGAAUCACCAACUGAACCAGCUCAAUCAGUUGAGCCACUUGAAUCAGUUCCAUCAGUUAAACCAAUUGAAUCAAUUGAGUCA ((((((......((..........)).(((........))).)))))).........((((((....))))))....(((.((((((...((((((.....))))))...)))))).))) ( -24.20) >DroSec_CAF1 5572 120 + 1 UGGUGAGACACAGCAUCACCUGCAGCAUGAACACCACCUGAAUCACCAACUGAACCAGCUCAAUCAGUUGAGCCACUUGAAUCAGUACCAUCAUUUAAGCCAAUUGAAUCAAUUGAGUAA ((((((..((..(((.....)))....((.....))..))..))))))(((((..((((((((....))))))....))..)))))............((((((((...)))))).)).. ( -24.80) >DroSim_CAF1 1065 120 + 1 UGGUGAGACACAGCAUCACCUGCAGCAUGAACACCACCUGAAUCACCAACUGAACCAGCUCAAUCAGUUGAGCCACUUGAAUCAGUACCAUCAUUUAAGCCAAUUGAAUCAAUUGAGUAA ((((((..((..(((.....)))....((.....))..))..))))))(((((..((((((((....))))))....))..)))))............((((((((...)))))).)).. ( -24.80) >DroYak_CAF1 1026 120 + 1 UGGUGAGCCACAGCAUCACCAUCAGCAUCAUCACCAUCUGAAUCACCAACUGAACCAGCUCAAUCAGUUGAGUCACUUGAAUCAGUUCCAUCAAUUAAAUCAAUUGAAUCAAUUGAGUCA ((((((((....)).))))))((((.((.......))))))...(((((((((..........))))))).))....(((.((((((...((((((.....))))))...)))))).))) ( -26.20) >consensus UGGUGAGACACAGCAUCACCUGCAGCAUCAACACCACCUGAAUCACCAACUGAACCAGCUCAAUCAGUUGAGCCACUUGAAUCAGUACCAUCAUUUAAACCAAUUGAAUCAAUUGAGUAA ((((((....(((........................)))..))))))(((((..((((((((....))))))....))..)))))............((((((((...)))))).)).. (-21.39 = -20.76 + -0.63)


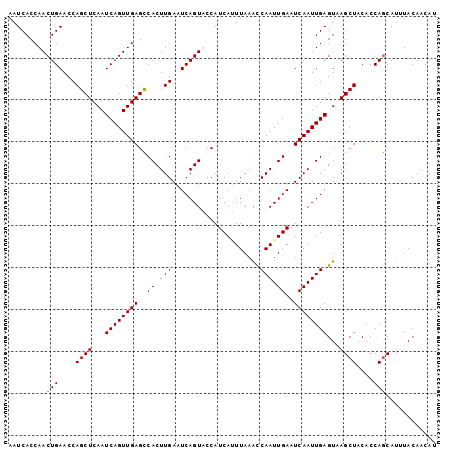
| Location | 4,114,402 – 4,114,504 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 94.44 |
| Mean single sequence MFE | -17.98 |
| Consensus MFE | -14.98 |
| Energy contribution | -15.22 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.923001 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4114402 102 + 22224390 AAUCACCAACUGAACCAGCUCAAUCAGUUGAGCCACUUGAAUCAGUUCCAUCAGUUAAACCAAUUGAAUCAAUUGAGUCAGCUACACCAGCAUAUACAACAU ......(((((((..........))))))).......(((.((((((...((((((.....))))))...)))))).)))(((.....)))........... ( -18.50) >DroSec_CAF1 5612 102 + 1 AAUCACCAACUGAACCAGCUCAAUCAGUUGAGCCACUUGAAUCAGUACCAUCAUUUAAGCCAAUUGAAUCAAUUGAGUAAGCUACACCAGCAUUUACAACAU .........(((....((((...((((((((..(((((((((.(......).))))))).....))..))))))))...))))....)))............ ( -17.60) >DroSim_CAF1 1105 102 + 1 AAUCACCAACUGAACCAGCUCAAUCAGUUGAGCCACUUGAAUCAGUACCAUCAUUUAAGCCAAUUGAAUCAAUUGAGUAAGCUACACCAGCAUUUACAACAU .........(((....((((...((((((((..(((((((((.(......).))))))).....))..))))))))...))))....)))............ ( -17.60) >DroYak_CAF1 1066 102 + 1 AAUCACCAACUGAACCAGCUCAAUCAGUUGAGUCACUUGAAUCAGUUCCAUCAAUUAAAUCAAUUGAAUCAAUUGAGUCAGCUACACCAACAUCUACACCAU ................((((...((((((((.(((.((((.(.((((.....)))).).)))).))).))))))))...))))................... ( -18.20) >consensus AAUCACCAACUGAACCAGCUCAAUCAGUUGAGCCACUUGAAUCAGUACCAUCAUUUAAACCAAUUGAAUCAAUUGAGUAAGCUACACCAGCAUUUACAACAU .........(((....((((...((((((((..((.(((.....................))).))..))))))))...))))....)))............ (-14.98 = -15.22 + 0.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:07:43 2006